Artificial Intelligence: The New Alexander Fleming
Article information
Almost a century after Alexander Fleming discovered penicillin and forever changed the face of medicine, Massachusetts Institute of Technology (MIT) researchers made another discovery potentially as valuable as Fleming’s ground-breaking work. They used artificial intelligence (AI) algorithms based on machine learning to trawl through a vast digital collection of pharmaceutical compounds to identify novel antibacterial molecules [1], and identified a new antibiotic that they dubbed halicin (named after 2001’s HAL 9000). Halicin, it turns out, acts in a manner different from conventional antimicrobials—by damaging the ability of bacteria to maintain an electrochemical gradient necessary for survival.
In the MIT study, a deep neural network model was developed to screen millions of molecules in mere days to identify potential antibiotics that employ mechanisms different from those of existing drugs. After screening numerous chemical libraries, the team identified halicin from the Drug Repurposing Hub [2], as a compound that shows bactericidal action against a wide assortment of pathogens, including Mycobacterium tuberculosis, carbapenem-resistant Enterobacteriaceae, Clostridioides difficile, and Acinetobacter baumannii. This seminal study underscores the value of AI for expanding our pharmaceutical arsenal to include structurally distinct antibacterial drugs.
Another potential role for AI in the realm of infectious diseases is to provide prescription guidance for antibiotics. The use of such decision support systems would be expected to reduce inappropriate prescriptions of antibiotics and antimicrobial resistance. Such systems would apply machine learning techniques or expert systems methods (Figure 1) to data from healthcare databases. Predictions based on a patient’s medical information would estimate the probability of the patient having a particular pathogen and provide an early warning regarding the risk of sepsis development [3].
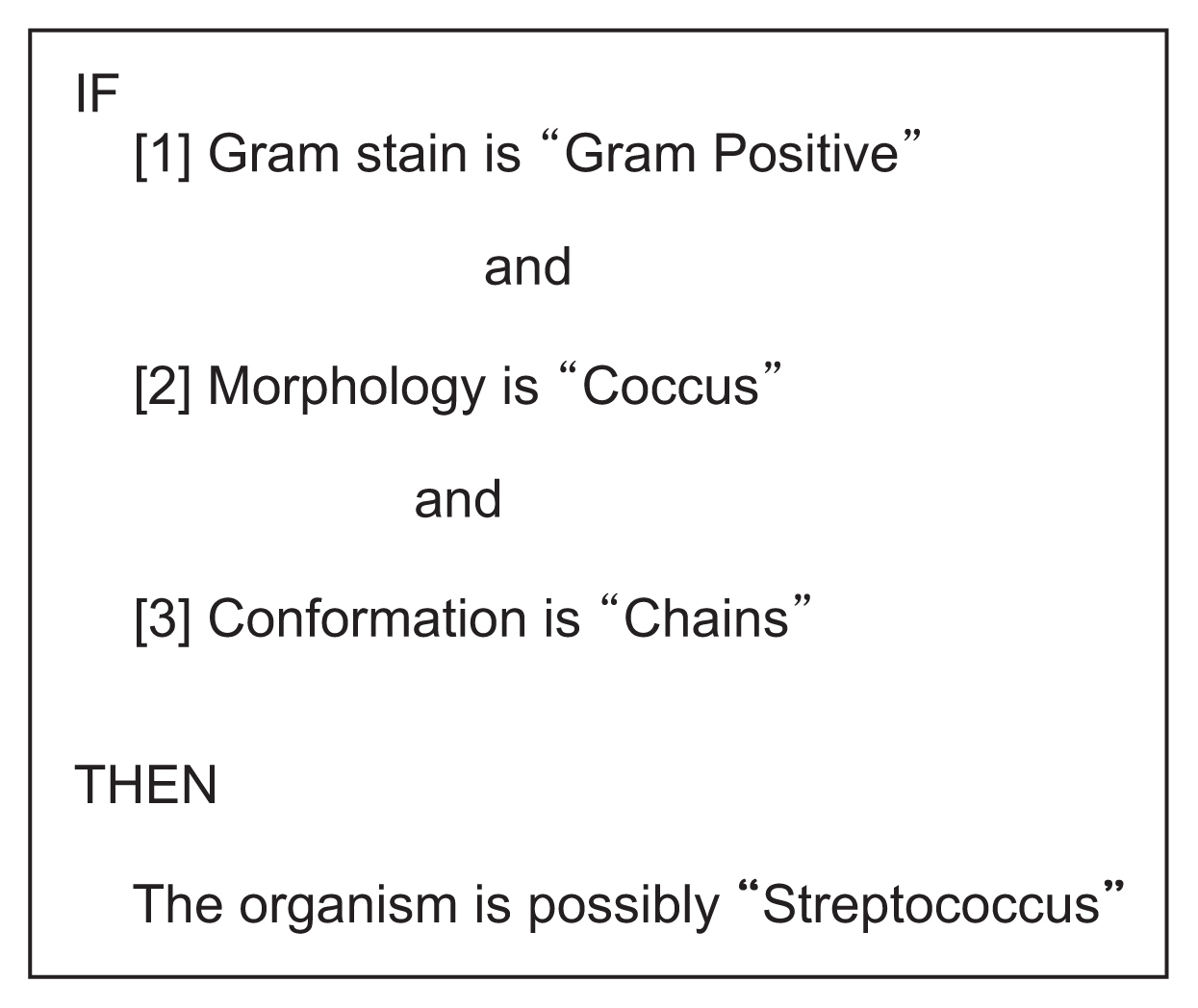
An example of an expert system rule pertaining to microbiology. This rule considers three organism-related factors (Gram stain, morphology, and conformation) to produce a conclusion that is probabilistic rather than certain. Example derived from Johnson L, Keravnou ET. Expert systems technology: a guide. Cambridge (MA): Abacus Press; 1985.
Excessive antibiotic use and the emergence of multi-drug resistant organisms (MDROs) is a major health problem [4,5]. Pathogens such as methicillin-resistant Staphylococcus aureus are now common and pose significant health risks. Clinicians are faced with a dilemma when treating sepsis caused by MDROs given the limited choice of effective antibiotics [6], as the indiscriminate use of antibiotics enhances the emergence of MDROs, which in turn burdens healthcare systems with the diagnosis and treatment of critically ill patients. This reality entails a hefty human and financial cost [7].
Antimicrobials work by disrupting bacterial multiplication, often by damaging the cell membrane (Table 1, Figure 2). However, few new antibiotics have been developed in recent decades, and there is a concern that we could eventually face the same problems that our ancestors experienced, with simple infections developing into life-threatening sepsis.

Illustration of some of the known mechanisms of antibiotic action. These include the inhibition of cell wall biosynthesis, inhibition of nucleic acid metabolism and repair, and protein synthesis inhibition, in addition to cell membrane disruption. Adapted from https://commons.wikimedia.org/wiki/File:Antibiotics_Mechanisms_of_action.png.
Antimicrobial resistance occurs through two principal mechanisms: mutation and the acquisition of resistance from another organism. In the former case, a mutated bacterium may (for example) produce an enzyme that inactivates a class of antibiotics or develop a mechanism to expel that antimicrobial from the bacterium before it reaches its cellular target. Known mechanisms of acquiring resistance from another organism include transfer of genetic information via conjugation, the acquisition of “naked” free DNA from the environment, or DNA transfer achieved via viral delivery [8].
Researchers have already collaborated on the use of AI in radiology, dermatology, pathology, and ophthalmology to improve care [9]. In addition to diagnosis, AI supports prognosis-related applications, such as predicting sepsis [10], with alerts generated by an AI model that provides an early warning. Such models predict deterioration and suggest possible pathogens and antibiotic susceptibility [11]. Schinkel et al. [12] reviewed the clinical applications of AI in sepsis and concluded that AI has the potential to prompt physicians when to intervene. Although these models offer a marginal advantage over current tools, data interpretation can be difficult [12]. Giacobbe et al. [13] reviewed applications of AI in the management of multidrug-resistant Gram-negative infections, classifying algorithms into applications such as predicting infection risk, identifying the aetiology, estimating the risk of emerging MDROs, and identifying antibiotic misuse [14].
The development of clinical decision support (CDS) systems remains a daunting challenge, as improving their positive predictive value is critical to improving performance and reducing alert fatigue. However, some studies may overestimate model performance due to poor adaptability and the definition of sepsis. There is also the matter of how alerts affect the provider’s actions and prescribing behaviour, as alert and CDS systems need to have a predictive value of over 60% to avoid alert fatigue [10]; therefore, developers need to understand these factors and their impact on clinical management. Unsurprisingly, evaluations of these systems by their own developers tend to report more favourable outcomes than evaluations conducted by a third party [15].
In conclusion, while medical AI offers promising opportunities to improve outcomes, our focus must be to solve clinical problems and not merely throw technology at patients. If history is kind to us again, in a hundred years, scientists will be looking back at us in the same way we viewed Alexander Fleming’s mould discovery.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
Zaki Almallah has been awarded a grant from Mubadala Healthcare for a proposed project on AI in antibiotics. No funding for writing this manuscript.

