 |
 |
- Search
| Healthc Inform Res > Volume 18(4); 2012 > Article |
Abstract
Objectives
Rare disease research requires a broad range of disease-related information for the discovery of causes of genetic disorders that are maladies caused by abnormalities in genes or chromosomes. A rarity in cases makes it difficult for researchers to elucidate definite inception. This knowledge base will be a major resource not only for clinicians, but also for the general public, who are unable to find consistent information on rare diseases in a single location.
Methods
We design a compact database schema for faster querying; its structure is optimized to store heterogeneous data sources. Then, clinicians at Seoul National University Hospital (SNUH) review and revise those resources. Additionally, we integrated other sources to capture genomic resources and clinical trials in detail on the Korean Rare Disease Knowledge base (KRDK).
Results
As a result, we have developed a Web-based knowledge base, KRDK, suitable for study of Mendelian diseases that commonly occur among Koreans. This knowledge base is comprised of disease summary and review, causal gene list, laboratory and clinic directory, patient registry, and so on. Furthermore, database for analyzing and giving access to human biological information and the clinical trial management system are integrated on KRDK.
Conclusions
We expect that KRDK, the first rare disease knowledge base in Korea, may contribute to collaborative research and be a reliable reference for application to clinical trials. Additionally, this knowledge base is ready for querying of drug information so that visitors can search a list of rare diseases that is relative to specific drugs. Visitors can have access to KRDK via http://www.snubi.org/software/raredisease/.
Certain definite phenotypes of rare diseases were actively researched among scientists to clarify its root even though a rare disease occurs infrequently in the total human population [1]. Accordingly, a few markers have been discovered and researchers have identified genetic origins [2]. Especially, the development of large-scale initiative in sequencing technologies has powerfully determined more rare variants in Mendelian disorders [3,4]. Many scientists are attracted to find uncovered elements in this field nowadays because they believe that it is the first step of approaching to cure rare chronic diseases.
Rare diseases are actually common despite of its rarity [5]. In other words, there are a large number of rare disease patients. According to EURORDIS, it is estimated that more than 5,000 distinct rare diseases exist and more than 6% of European are affected by rare diseases. In addition, there might be a number of unreported cases, ultra-rare diseases, and few patients that do not have obvious characteristic or symptom [6]. In some populations, prevalence of rare diseases is dramatically high due to genetic inheritance. Hence, academics try to take a look into the ethnic characteristics in order to identify a clue of genetic disease which mainly occurs between populations [7].
Nevertheless, Korean researchers refer foreign resources to get information for rare diseases. Applying new guideline of treatments from different ethnic group would be inappropriate for Korean patients. A great number of health care providers and consumers are seeking for valuable information [8], however, some materials online does not help for domestic researchers such as: laboratory and clinic directory. It is necessary to have data interchange hub for national so that researchers would have meaningful data for rare disease that mainly occurs among Korean.
According to Centers for Disease Control (CDC), approximately 500,000 of people are suffering from more than 110 kinds of rare diseases in Korea. Strictly speaking, approximately 0.1% of Korean population has been attacked by a few diseases and most of them have not started any research yet. Moreover, it is hard to find resources-genetic counseling, disease treatment, care center information-for sufferers and their family. The only way to get information is consulting a doctor.
Therefore, we have developed Korean rare disease knowledge base. Its aim is to contribute to the collaboration work between practitioners who are interested in the same subjects for better results. Rare disease knowledge base can provide overall tips to subjects for better understanding and treatment. Additionally, the first step of this approach will make people to begin paying attention to the problems of carelessness with orphan disease. The knowledge base is comprised of disease summary, review article, genetic variation, laboratory and clinic directory, patient registry, and domestic research. Although drug database is not yet ready in Korean Rare Disease Knowledge base (KRDK), it is already considered for storing list of drug that associated with genes when we design database schema. For the last step, we also developed a Web-base interface with user friendly to enable to search and find knowledge instantly with a few clicks.
GeneTests is a database developed at University of Washington for free online medical genetics information resources (http://www.genetests.org) [9]. We referred its schema for designing a new one and made an optimized database schema for storing information about summary of disease, review, genetic variant, laboratory and clinic directory, patient registry, and domestic research on KRDK. It helps to execute querying quickly and avoid redundant data. Additionally, we considered storing drug information that is highly connected to target gene and disease. As a result, extended version of database schema is ready for disease, gene, and drug relations and its relation based query is executable (Figure 1).
Orphanet, which runs by a consortium of European partners, is a database of information on rare diseases and orphan drugs for all public (http://www.orpha.net) [10]. It provides overall information about disease and its genetic data. 8 practitioners in Seoul National University Hospital (SNUH) generally referred and revised Orphanet review articles and other public databases, then they arrange a set of structured-revision data and we stored over 500 disease summaries and 48 reviews which are translated in Korean. We collected national laboratory and clinic directory by contacting experts and directors. Once data supplied voluntarily, Research Center for Rare Diseases (RCRD, http://rarediseasecenter.org) confirmed and guaranteed those data contain laboratory and clinic name, director's name, contacts and detail information. Specifically, lab directors provide molecular genetic testing for a specific disorder. Finally, we have attempted to make the listing of Korean clinical laboratory comprehensive and stored reliable information in order to offer directory lists by searching specific disease.
Bio Electronic Medical Record (BioEMR) is clinical trials management system (http://bioemr.snubi.org:8080/bioemr/rdrc/) that was developed at Seoul National University Biomedical Informatics (SNUBI) [11]. Patient's records in BioEMR, however, are not open access data due to private information. Accordingly, we summed up and offer only summary of patient's registry of each disease. It is also available to have access registry data in detail by contacting RCRD. Genome Research Information Pipeline (GRIP) is an integrated database for analyzing and having access biological information of human, mouse, and rat (http://grip.snubi.org). GRIP consists of a number of major biological databases and contains information about sequence, gene, protein, gene family, protein family, and enzyme. We integrated the KRDK with GRIP to capture the all biological information related the gene name or symbol.
KRDK contains 520 rare disease entries and 48 disease reviews. In addition, 6 laboratory directories include testing methods on 303 inherited diseases for 184 genes and 35 specialists in rare diseases in Korea are listed as well. There are 839 discovered genes that are known to affect rare diseases and its chromosomal location and protein name are also provided. It contains 12 disease patients' registries and 22 ongoing domestic researches with study title, director contact, and so on (Table 1).
User may search specific disease with its name-it does not need to be a full name of disease-in either Korean or English. KRDK can be searchable by disease name, gene symbol, protein name in order to explore a certain rare disease and disease name can be used to find related laboratory and clinic directory. On the other hand, users may have a look of whole registered rare diseases by clicking 'All Disease' button at the bottom.
Every search result is displayed alphabetical order for all diseases. KRDK is comprised of well defined categories with highly structured sections (Figure 2). For every single disease, a search result shows 5 buttons (summary, testing, clinic, registry, and research) and two different colors (blue signifies activated button and gray signifies inactivated button) of button represents availability of its content.
Summary section provides rare disease name, prevalence, inheritance, age of onset, the International Statistical Classification of Diseases (ICD) 10 codes, Mendelian Inheritance in Man (MIM) number, disease synonym, and a summarized article of disease in Korean. Disease related genetic information are shown on the same page and provide gene symbol, chromosomal location and protein name. Additionally, every MIM number has hyperlink to online MIM (OMIM) Website and gene symbol links to GRIP for more information related to a specific gene. Each of review includes definition, prevalence, mechanism, diagnosis, treatment, genetic counseling and prenatal diagnosis. Those reviews were referred to previous researches and written by consultant of each specific disease.
Above all, laboratory and clinic directory are domestic information. Laboratory directory focuses on testing usage in diagnosis and target genes for a specific disease. Also, its contact information has informed (Figure 3A). On the other hand, clinic directory provides not only a clinic name but also a list of name of a practitioner who is a specialist of a specific rare disease and contacts (Figure 3B).
BioEMR is a patient registry developed at SNUBI and it is not accessible database for public because of patient information. Consequently, we provide information about the name of registry, the number of registered patients, the number of registered genes, a name of director, contact and BioEMR URL so that scientist who wants to get patient reports in detail can request data for referring each record of patients (Figure 4).
Lastly, we investigated and stored summary of rare disease national studies. This section includes study title, institute, subject related references, director's name and contact. A clinician who is interested in a specific rare disorder will have a chance to cooperate for the better research outcome. On the other hand, it is good to avoid two similar experimentations in different groups. With the many domestic researches, more and more people will pay attention to concern genetic disorders.
A unified format makes it easy for data handler to collect data without information loss and to keep data consistency. However, each of institute has its own format for recording. Each of outlines needs to be modified depending on purpose of usage. Hence, we have been offering a Web-based online submission tool with a single design for updating and modifying investigated information and test results without cost. Only experts may submit knowledge so there is a restriction-it requires sign in process-on having access to submission tool because untested information would mislead beginner practitioners and it is all about a clinical matter of life and death.
This paper introduces the first rare disease knowledge base for Korean. It is comprised of comprehensive rare disease, genetic variants, disease review, laboratory directory for molecular genetic testing, clinic directory for diagnosis and care, patient's registry, and ongoing rare disease study. Such resources are spread out online, therefore, we developed rare disease knowledge base integrated with patient's registry database and database for having access biological information of human. Yet there is not enough data for those who are interested in rare disease, we expect that the number of input data will exponentially increase sooner or later.
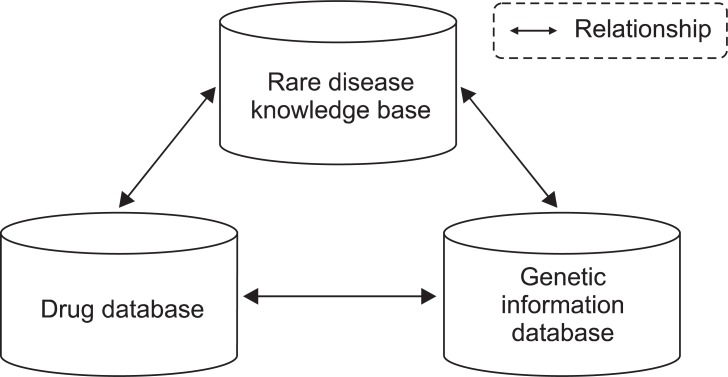
KRDK integrate other sources like GRIP and BioEMR in order to provide comprehensive disease information. Consequently, physicians do not need to spend more time on gathering patient's record and referring other biological databases for more information. By clicking gene symbol, visitors will instantly get information about gene family, protein, protein family, enzyme, and so on. It is also able to store drug information and each drug has relationships between drug, target gene and disease. Performing query with drug names shows disease and target gene list, and vice versa (Figure 5). Though we focused on gathering rare disease sources on KRDK, we have primarily concerned about data reliability. Experts double checked all disease summary and reviews in order to provide accurate knowledge. Hence, not only physicians but also patients who may not know much in rare diseases will get useful and practical information from this knowledge base.
Online submission tool helps to collect more information easily from various resources, therefore, submitted materials need to be confirmed by experts. It is necessary to make a group of experts on rare disease for verifying given data to offer reliable information. With amount of credible data, we expect that this knowledge base contribute to help collaborative research and apply for clinical trials. An effort by creating Korean knowledge base for improving understanding on rare diseases among Korean is the most valuable infrastructure in research field.
More and more variants were identified by next generation sequencing technology [12,13]. There is great interest in investigating whole-exome sequencing data for deciphering rare variants which can be a key role in the etiology of rare disease. The price is affordable for exon and so is whole-genome sequencing recently. Affecting variants to rare disease can be distinguished by trio or quartet sequencing among family. We assume that discovered variants also can be alarmed in the same race and there would be more and more identified causal variants from sequencing data analysis. Consequently, we are considering that building a database for genetic variants with exome and whole-genome sequencing data so that practitioners or patients can refer their own sequenced data on KRDK.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012-0000994).
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A080588 and A120017).
References
2. Pescucci C, Mari F, Longo I, Vogiatzi P, Caselli R, Scala E, et al. Autosomal-dominant Alport syndrome: natural history of a disease due to COL4A3 or COL4A4 gene. Kidney Int 2004;65(5):1598-1603. PMID: 15086897.


3. Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A 2009;106(45):19096-19101. PMID: 19861545.



4. Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 2010;42(1):30-35. PMID: 19915526.


5. EURORDIS. Rare diseases: understanding this public health priority [Internet]. 2005. cited at 2012 Dec 1. Paris: EURORDIS; Available from: http://www.eurordis.org/IMG/pdf/princeps_document-EN.pdf

6. Morgan SS, Aslam MB, Mukkanna KS, Ampat G. A rare presentation of sarcoidosis, back pain and spondylolisthesis. J Bone Joint Surg Br 2008;90(2):240-242. PMID: 18256097.


7. Palo JU, Ulmanen I, Lukka M, Ellonen P, Sajantila A. Genetic markers and population history: Finland revisited. Eur J Hum Genet 2009;17(10):1336-1346. PMID: 19367325.



8. Pagon RA, Tarczy-Hornoch P, Baskin PK, Edwards JE, Covington ML, Espeseth M, et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat 2002;19(5):501-509. PMID: 11968082.


10. Weinreich SS, Mangon R, Sikkens JJ, Teeuw ME, Cornel MC. Orphanet: a European database for rare diseases. Ned Tijdschr Geneeskd 2008;152(9):518-519. PMID: 18389888.


11. Park YR, Bae YJ, Kim JH. BioEMR: an integrative framework for cancer research with multiple genomic technologies. Summit on Translat Bioinforma 2008;2008:81-84. PMID: 21347128.



12. Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009;461(7261):272-276. PMID: 19684571.



13. Erlich Y, Edvardson S, Hodges E, Zenvirt S, Thekkat P, Shaag A, et al. Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res 2011;21(5):658-664. PMID: 21487076.



Figure 1
An optimized database schema for Korean Rare Disease Knowledge base (KRDK). Well-structured database keeps data integrity, avoiding data redundancy problem, and appropriately organized data structure helps searching effectively.
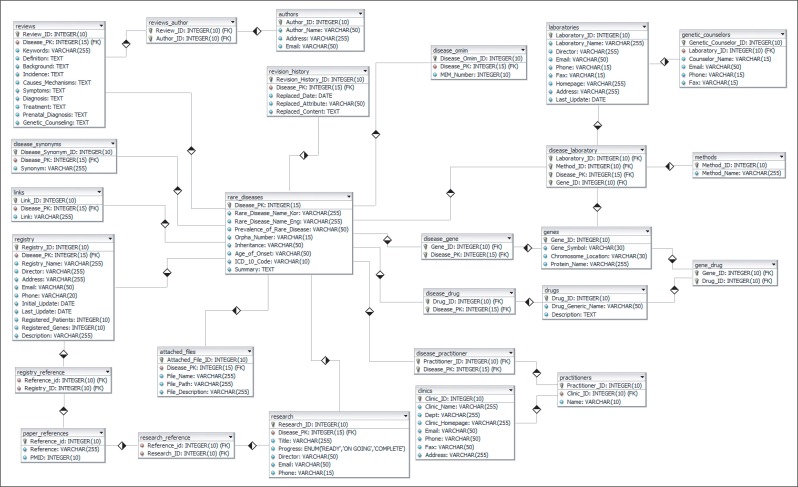
Figure 2
The search result page of disease and additional information. Summary section shows overall disease information briefly. With clicking gene symbol, it directly connects to Genome Research Information Pipeline (GRIP) for more information that relates to a specific gene such as: protein, protein family, pathway and so on. Moreover, Mendelian Inheritance in Man (MIM) number have hyperlink to online MIM (OMIM) so that user may browse on Website instantly.

Figure 3
An example of laboratory and clinic directory pages. Search result for laboratory and clinic directory pages. (A) All laboratories are list with available testing method with target gene. Additionally, by clicking laboratory name, visitors may get its information in detail such location, contact. (B) Clinic directory shows a name of practitioner that is a specialist of a specific disease and other information like clinic name and contact.

Figure 4
The summary of patient's registry and its original Web page link. Patient's registry shows a summary of records due to privacy policy. Summary is comprised of the number of registered patient and gene, director, contact, references and so on. Additionally, patient's registry URL directly links and visitors can fully have access to permission from Research Center for Rare Diseases (RCRD).

Figure 5
Drug information that is highly connected to rare disease and genetic information database. We are ready for storing drug information on Korean Rare Disease Knowledge base (KRDK). Drugs are highly connected to target genes and specific diseases. With this relationship, physician can perform several possible queries; What are drugs relative to Wilson disease? What target genes do Penicillamine have? What are genes that affect Wilson disease? By answering those queries, clinician can analyze research efficiently with this integrative database scheme.
-
METRICS

- Related articles in Healthc Inform Res
-
The Development of a Korean Drug Dosing Database2011 December;17(4)
Development of Korean Oriental Nursing Terminology Set2007 June;13(2)
Development of an Evaluation Checklist for Internet Health/Disease Information2006 December;12(4)
Development of a PDA based Personal Asthma Management System2005 September;11(3)






