|
 |
- Search
| Healthc Inform Res > Volume 18(3); 2012 > Article |
Abstract
Objectives
To develop dedicated markup language for clinical contents models (CCM) to facilitate the active use of CCM in electronic health record systems.
Methods
Based on analysis of the structure and characteristics of CCM in the clinical domain, we designed extensible markup language (XML) based CCM markup language (CCML) schema manually.
With the increase in aging population and chronic disease patients, the interest of clinical information systems supporting coordination and continuity of care have been rising. Lifelong Electronic Health Record (EHR) systems have been a popular research agenda in many countries [1-3]. To support coordination and continuity of care, clinical information of patient within EHR systems should be represented in a standardized way to ensure interoperability [4-6], and able to capture detailed clinical information to support clinical decision [7].
The detailed clinical model (DCM) is an information models which support interoperability and detailed capture of EHR data by standardized representation of clinical information [8,9]. For the active use of DCM, a markup language for DCM is a prerequisite because it allows computer systems to process DCM electronically. The followings are examples of DCMs and their markup languages for the formalisms: clinical element model and clinical element model language (CEML) at Intermountain Healthcare [10], Health Level 7 (HL7) template and HL7 V3 XML by HL7 International [11], archetypes and archetypes definition language (ADL) and resource description framework (RDF) defined by the openEHR Foundation [12], and the clinical information model and unified modeling language (UML) and XML in the Netherlands [13].
Since 2007, the Center for Interoperable EHR [14] has developed clinical contents model (CCM) which is a type of DCM incorporating definitions of the essential attributes and values of the health information to be observed and recorded in patient care [15]. The goal of CCM is to support interoperability of the EHR data for various sectors and jurisdictions. To meet this goal, the design of the CCM was based on ISO 18308 EHR requirement [16] and ISO 20514 EHR scope [17] to guarantee detailed capture and effective retrieval of EHR data.
To facilitate active use of CCM within EHR systems, CCM should be expressed in machine readable format for computer systems to electronically process them. However, computer systems could not process CCM with its original format because it is a knowledge content expressed in Microsoft excel format. Therefore, we developed CCM markup language (CCML) that reflects the structure and characteristics of CCM accurately. Use of CCML can ensure electronic processing of CCM in computer systems. To guarantee technology neutrality and utilization without any special tool or system, it is based on XML.
This study developed CCML by analyzing around 2,200 cases of CCM developed so far in the clinical observation domain. As CCM in the medication and laboratory domains are still being refined, they were excluded from this study. Based on analysis of the structure and characteristics of CCM, the schema of CCML was designed and developed manually by the authors who were experts in CCM development.
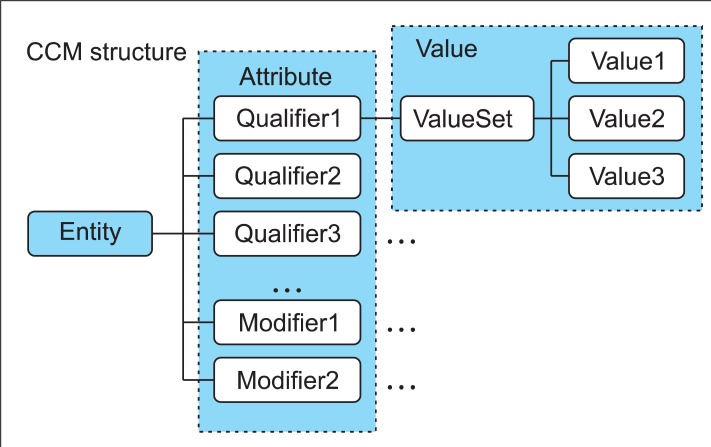
As in Figure 1, CCM is expressed in the structure of Entity, Attribute (Qualifiers/Modifier), and Value. A model has one Entity in principle but, if necessary, may have two or more Entities. Each Entity is an independent clinical concept and has one or more Attributes for representing the attributes of the clinical concept. Attributes are largely divided into Qualifiers for representing the attributes of the clinical concept and Modifiers for changing the attribute values of the clinical concept. The specific value expressed by an Attribute is contained in Value.
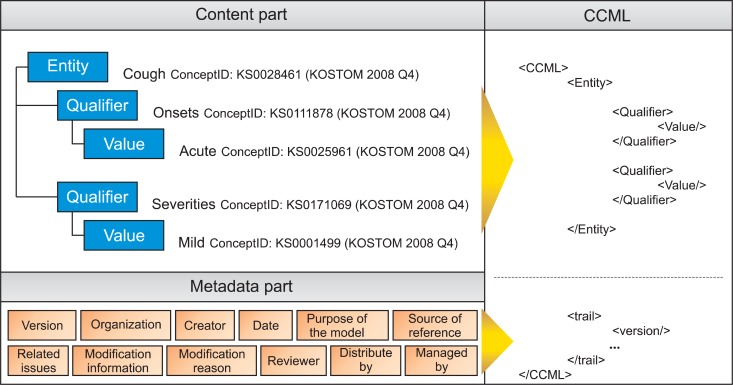
For example, a CCM model for cough is as follows. The model is "CoughAssert" and the Entity of the model is "cough." In the entity, the onset of the sickness is expressed by Qualifier "dateOfOnset" and the degree of it is expressed by Qualifier "severity." For Qualifier "severity," the ValueSet includes "mild," "mild to moderate," "moderate to severe," "severe." When patient information is entered into an actual computer system, one of values in the value set is selected.
Modifiers include subjectOfInformation, negationInd, and uncertainty. The subjectOfInformation is a subject of CCM Entity. Default value is patient, but it can also be fetus, organ donor, or informant. The negationInd is for negating the entity. For example, instead of making a model for expressing "the absence of hypertension symptoms," we can express the absence of hypertension symptoms by setting the value of negationInd to "Yes" in the "hypertension" model. The uncertainty is for uncertain information. It has the value of "Yes" if symptoms are uncertain.
In order for such clinical concepts to be used as computer-based information, CCM has attributes as an information model. That is, it has information such as model type, clinical domain, cardinality, data type and Korean Standard Terminology of Medicine (KOSTOM) [18] for mapping to standard health terminology systems [19]. Model type indicates whether a model is an atomic model with one entity or a compound model with multiple entities, and clinical domain indicates clinical domain such as clincal finding, medication order, laboratory observation. Cardinality is the appearance frequency of a Qualifier or a Value. Data types used in CCM are defined according to HL7 V3 data type [20,21].
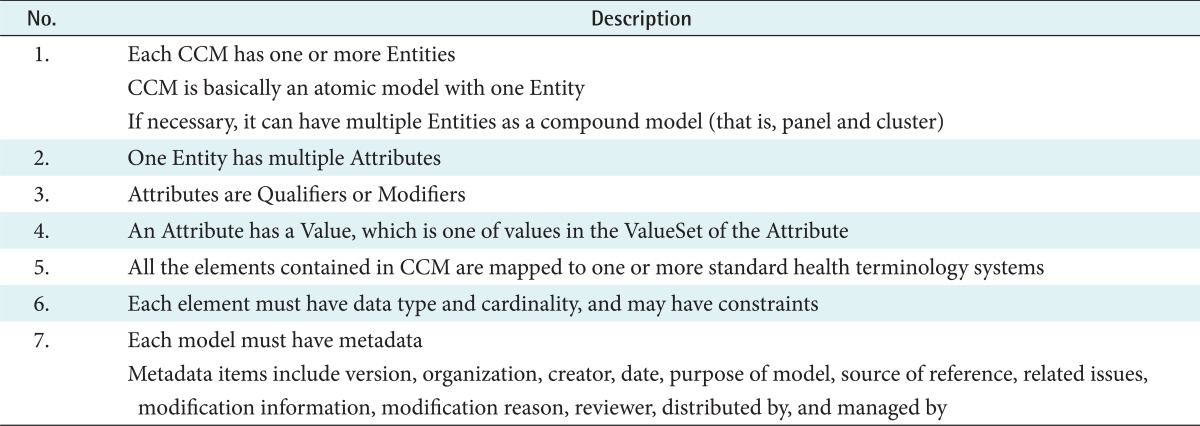
In addition to information for expressing a clinical concept, CCM has metadata for the development and management of the model. Metadata includes version, development institution, model developer, date of generation, purpose of model development, references for model development, related issues, information on change, reasons for change, reviewers, distribution institution, and management institution. Using metadata, the model developer can focus on the contents development, and the model manager can manage the change of the model. Model users can learn how to use the model accurately from the purpose of development and references used in model development.
Table 1 shows the characteristics of CCM recognized through analyzing its structure.
CCM has a tree structure. An Entity has multiple Qualifiers and Modifiers, and each Qualifier and Modifier has their own Values. The top node is Entity, and the other CCM components including Qualifiers, Modifiers, and Values are child node of it. This structure is well fitted XML. Furthermore, XML is highly readable and technology-neutral, so that it can be usable without additional efforts such as the development of a separate parser. Given these advantages, CCML was developed on XML-based.
From the tree structure of CCM, Figure 2 shows the schematic structure of XML-based CCML. As in Figure 2, the Entity, Qualifiers, Values and metadata information of CCM are converted easily to XML structure. The syntax of CCML elements uses keywords in CCM such as 〈CCML〉, 〈Entity〉, 〈Qualifier〉, and 〈Value〉 as they are, and metadata includes 〈version〉, 〈organization〉, 〈creator〉, etc., using the 〈trail〉 element. As metadata is used by the model developer, manager and users only for understanding situations surrounding the model, it may be deleted in constructing a system using the model. Therefore, metadata is expressed in the 〈trail〉 element. Metadata contained in a CCM includes all information about the changes of the model from ver. 0.1 to 1.0, so it occupies a very large size in the CCML file. Therefore, it is considered reasonable to delete 〈trail〉 information when an actual system is constructed based on CCM.
The Characteristics of CCM as an information model is expressed as child elements or attributes. It means that the information such as data type, cardinality and mapping information to standard health terminology systems is converted to child elements or attributes. Details are described in the next chapter.
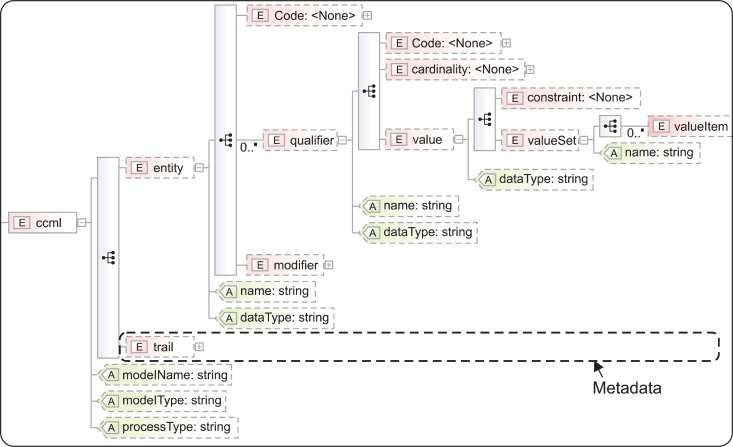
Reflecting the structural characteristics of CCM in Table 1 and metadata, we generated a schema manually as in Figure 3. The root element is 〈ccml〉, and it has attributes model name, model type, and process type. Entity 〈entity〉 has attributes name and data type, and information on standard health terminology systems mapped to the entity is expressed in the attributes of element 〈Code〉, which are code, codeSystem, CodeSystemName, and displayName. 〈qualifier〉 also has attributes name and data type, and child elements 〈Code〉, 〈cardinality〉 and 〈value〉. Metadata is expressed in 〈trail〉. According to episode of metadata generation, 〈trail〉 has 〈version〉, 〈organization〉, 〈creator〉, 〈date〉, 〈purposeOfModel〉, 〈sourceOfReference〉, 〈relatedIssues〉, 〈modificationInformation〉, 〈modificationReason〉, 〈reviewer〉, 〈distributedBy〉, and 〈managedBy〉.
Figure 4 is an exemplary display of model AbdominalPainAssert that the CCM manager can view. According to the contents, "AbdominalPainAssert" and "AbdominalPain" have qualifiers dateOfOnset, duration, lasting, frequency, etc., and qualifier "periodOfOnset" can have a value of "Acute" or "Subacute."
Figure 5 shows metadata for model AbdominalPainAssert. The content part of the model specifies the contents of the latest version in CCML, but metadata adds the history of changes continuously as separate episodes. In the example of Figure 5, version information is v1.0, which indicates that the model is the final version completed, and episode number 8 shows that the model has been changed 8 times. In the CCM file of model AbdominalPainAssert, the 〈trail〉 element includes all the contents from 〈episode number="1"〉 to 〈episode number="8"〉. Thus, the history of the model development can be traced back. It was confirmed by model developers including several clinicians that all information to be expressed in CCM is represented well in CCML using XML. We converted around 2,200 developed CCM products into CCML and published the results in CCM Manager (http://www.clinicalcontentsmodel.org), the website for advertising CCM products.
Figures 4 and 5 show only an atomic model, but the CCM manager site provides compound models such as Cluster and Panel. Total 12 Cluster models were developed including AffectCluster, CerebellarSignCluster, ChestAuscultationCluster, and VitalSignCluster. Also total 12 Panel models were provided including ApgarScorePanel, BloodPressurePanel, and GlasgowComaScalePanel.
CCML enables the composition of clinical document templates or structured data entries (SDE) by expressing CCM in a XML-based patterned structure. Each CCM model can be a component forming a section of a clinical document. Qualifiers and ValueSets contained in a CCM model are presented as combo boxes or radio buttons, and by clicking or selecting them, users can enter clinical information easily and accurately.
Qualifier "〈periodOfOnset〉" in Figure 4 has value "Acute" or "Subacute," and the value can be accessed through XPATH, and presented as combo boxes or radio buttons on the screen using JavaScript. However, CCML alone is not enough for clinicians to enter data fast and accurately using CCM. It should be supported by customizing in consideration of each hospital's environment and work process.
In this paper we developed a CCML for the active use of CCM in EHR systems. CCML is a highly human-readable and technology-neutral markup language that supports the representation of the unique characteristics and structure of CCM as they are. As the developed CCML expresses in XML, it can be introduced to a system and processed in the system readily using various XML technologies including XSTL, XQuery, and XPath, without any special device or technological support.
The advanced health information models such as HL7 V3 RIM, openEHR's Archetype and CEM have their own model languages-Clinical Document Architecture (CDA), ADL, and CEML. Compared to CDA, ADL, and CEML, Table 2 shows the comparison result.
First, CCML, CDA, and ADL are all machine readable. The basis whether the model is machine readable or not is to provide coded-information not just narrative text. It means that machine require the information to be interpreted without character recognition technology. Therefore, CCML, CDA, and ADL compose the contents with information mapping into standard health terminology. However, CEML do not contain mapping information inside model. It just has links to data dictionaries physically outside the model [22].
CDA's entry leveled body is composed of 9 clinical statements classes so it is very powerful to express machine readable contents. However, it is highly recursive and nested structure, so it is very difficult for human to interpret the data. Even if it is machine, it requires a lot of efforts to visualize and process data. In comparison, CCML presents information in a much more intuitive way than CDA. Because it expresses the structure of CCM directly, it is less completed. The simplicity reduces the efforts to visualize and process the internal data of model. Although CCML is simpler than CDA, every CCM component is mapped into standard health terminology system so that all contents are machine readable.
Secondly, CCML, CDA, and CEML are based on XML, so they can be processed in technical neutral way. ADL has an excellent structure to express complicated clinical concept accurately, it uses its unique grammar and syntax. Therefore it requires additional effort to develop the more advanced agnostic parser. As CCML was developed through the specialization of CCM, it should continuously reflect changes of CCM such as modification of structure or newly added characteristics of it. By comparison the other model language, the results shows that 1) CCML presents the clinical concept in easy way, 2) it do not require any other effort to develop or buy a parser, and 3) it can be useful in EHR systems if the developing of CCM will be done.
The utility of CCML was demonstrated in Sepsis Management Pilot System (SMPS) in the National Police Hospital which is a clinical decision supporting systems (CDSS) built on CCM for sepsis management in 2010 [23]. The SMPS provided clear evidence that CCML can fully support functions of CCM in an EHR system. The goal of CCML is to support the exchange and reuse of health information across various computer systems through active use of CCM. To achieve this goal, CCM and CCML should meet the requirement of ISO 18308 EHR Requirement which is technical specification contains a set of clinical and technical requirements for EMR architecture. CCM data type conform HL7 V3 XML data type. This logical information model supports communication and representation of clinical information which follow ISO 20514. We expect more active development of various application programs based on CCML such structured data entries system and CDSS in EHR systems.
Acknowledgments
This study was supported by a grant of the Korea Health 21 R&D Project, Ministry of Health & Welfare in Republic of Korea (A050909).
References
1. Heitmann KU, Schweiger R, Dudeck J. Discharge and referral data exchange using global standards: the SCIPHOX project in Germany. Int J Med Inform 2003;70(2-3):195-203. PMID: 12909170.


2. Integrating the Healthcare Enterprise. IHE laboratory technical framework supplement 2006-2007 [Internet]. 2006. cited at 2012 Sep 15. Integrating the Healthcare Enterprise; Sharing laboratory report (XD*-LAB). Available from: http://www.ihe.net/Technical_Framework/upload/IHE_LAB_TF_Sharing_Lab_Report_XD_Lab_TI_2006_09_14.pdf

3. Health Level Seven International. Continuity of care document [Internet]. c2012. cited at 2012 Sep 1. Ann Arbor: HL7; Available from: http://www.hl7.org/documentcenter/public_temp_D458FA83-1C23-BA17-0CB3B858E7E4DC44/pressreleases/20070212.pdf

4. Braun LM, Wiesman F, van den Herik HJ, Hasman A, Korsten E. Towards patient-related information needs. Int J Med Inform 2007;76(2-3):246-251. PMID: 16647878.


5. Kwak YS. Electronic health record: definition, categories and standards. J Korean Soc Med Inform 2005;11(1):1-15.

6. Haux R. Health information systems - past, present, future. Int J Med Inform 2006;75(3-4):268-281. PMID: 16169771.


7. Rector AL, Nowlan WA, Kay S. Foundations for an electronic medical record. Methods Inf Med 1991;30(3):179-186. PMID: 1943789.


8. Huff SM, Rocha RA, Bray BE, Warner HR, Haug PJ. An event model of medical information representation. J Am Med Inform Assoc 1995;2(2):116-134. PMID: 7743315.



9. Goossen W, Goossen-Baremans A, van der Zel M. Detailed clinical models: a review. Healthc Inform Res 2010;16(4):201-214. PMID: 21818440.



10. Parker CG, Rocha RA, Campbell JR, Tu SW, Huff SM. Detailed clinical models for sharable, executable guidelines. Stud Health Technol Inform 2004;107(Pt 1):145-148. PMID: 15360792.


11. Dolin RH, Alschuler L, Beebe C, Biron PV, Boyer SL, Essin D, et al. The HL7 clinical document architecture. J Am Med Inform Assoc 2001;8(6):552-569. PMID: 11687563.



12. Garde S, Chen R, Leslie H, Beale T, McNicoll I, Heard S. Archetype-based knowledge management for semantic interoperability of electronic health records. Stud Health Technol Inform 2009;150:1007-1011. PMID: 19745465.


13. Goossen WT. Using detailed clinical models to bridge the gap between clinicians and HIT. Stud Health Technol Inform 2008;141:3-10. PMID: 18953119.


14. Center for Interoperable HER [Internet]. c2012. cited at 2012 Sep 1. Seoul: CiEHR; Available from: http://www.ehrkorea.org/

15. Ahn SJ, Kim Y, Yun JH, Ryu S, Cho K, Kim S, et al. Clinical contents model to ensure semantic interoperability of clinical information. J KIISE Softw Appl 2010;37(12):871-881.

16. International Organization for Standardization. Health informatics: requirement for an electronic health record architecture. 2011. Geneva, Switzerland: International Organization for Standardization; (ISO 18308:2011)

17. International Organization for Standardization. Health informatics: electronic health record-definition, scope and context. 2005. Geneva, Switzerland: International Organization for Standardization; (ISO/TR 20514:2005)

18. Korea Standard Terminology of Medicine: KOSTOM [Internet]. c2012. cited at 2012 Sep 1. Seoul: Korea Health and Welfare Information Service; Available from: http://www.khwis.or.kr/

19. Ahn SJ. Development and application of development principles for clinical information model. J Korea Acad Industr Coop Soc 2010;11:2899-2905.

20. Dolin RH, Alschuler L, Boyer S, Beebe C, Behlen FM, Biron PV, et al. HL7 Clinical Document Architecture, Release 2. J Am Med Inform Assoc 2006;13(1):30-39. PMID: 16221939.



21. Beeler GW. HL7 version 3: an object-oriented methodology for collaborative standards development. Int J Med Inform 1998;48(1-3):151-161. PMID: 9600415.


22. Coyle JF, Mori AR, Huff SM. Standards for detailed clinical models as the basis for medical data exchange and decision support. Int J Med Inform 2003;69(2-3):157-174. PMID: 12810120.


-
METRICS

- Related articles in Healthc Inform Res
-
Prevalence of Sharing Access Credentials in Electronic Medical Records2017 July;23(3)
Conformance Analysis of Clinical Pathway Using Electronic Health Record Data2015 July;21(3)