Medical Image Retrieval: Past and Present
Article information
Abstract
With the widespread dissemination of picture archiving and communication systems (PACSs) in hospitals, the amount of imaging data is rapidly increasing. Effective image retrieval systems are required to manage these complex and large image databases. The authors reviewed the past development and the present state of medical image retrieval systems including text-based and content-based systems. In order to provide a more effective image retrieval service, the intelligent content-based retrieval systems combined with semantic systems are required.
I. Introduction
With the widespread use of picture archiving and communication system (PACS) in the hospitals, the amount of medical image data is rapidly increasing. Thus, the more efficient and effective retrieval methods are required for better management of medical image information.
There are two ways that medical images are retrieved, text-based and content-based methods [1]. In text-based image retrieval, the images are retrieved by the manually annotated text descriptions and traditional database techniques to manage images are used. In content-based image retrieval, the images are retrieved on the basis of features such as color, texture, shape, and so on, which were derived from the images themselves.
So far, a variety of medical image retrieval systems have been developed using either method (text-based or content-based) or combining two methods; a rough classification of medical image retrieval methods is displayed in Figure 1.
Therefore, we described two technological approaches for medical image retrieval - past development and present status, and future perspectives.
II. Text-Based Medical Image Retrieval
Text-based image retrieval system can be traced back to 1970s. Text-based image retrieval system is prevalent in the search on the internet web browsers. Although text-based methods are fast and reliable when images are well annotated, they cannot search in unannotated image databases. Moreover, text-based image retrieval has the following additional drawbacks, it requires time-consuming annotation procedures and the annotation is subjective [2]. Text-based query commonly results in irrelevant images (Figure 2). Thus, to support effective image searching, retrieval methods based on the image content were developed.

Text-based query commonly retrieves irrelevant images on the internet web browser. For user query "bike (bicycle)," irrelevantly retrieved images (not relevant to bike) are shown because image annotations contain a word "bike" such as bike tour, school bike rack, water bike, abbreviation, etc. (upper two layers). Miscellaneous medical images retrieved by query of "Reconstruction and computed tomography (CT)" are displayed in the lower 3rd layer of the figure (CT for image reconstruction, 3-dimensional reconstruction CT image, CT reconstruction algorithm, bone reconstruction with CT, breast reconstruction using CT).
III. Content-Based Medical Image Retrieval
In content-based image retrieval systems, images are indexed and retrieved from databases based on their visual content (image features) such as color, texture, shape, etc. Commercial content-based image retrieval systems have been developed, such as QBIC [3], Photobook [4], Virage [5], VisualSEEK [6], Netra [7]. Eakins [8] has divided these image features into three levels as followings: 1) Level 1 - Primitive features such as color, texture, shape or the spatial location of image elements. Typical query example is 'find pictures like this'; 2) Level 2 - Derived attributes or logical features, involving some degree of inference about the identity of the objects depicted in the image. Typical query example is 'find a picture of a flower'; 3) Level 3 - Abstract attributes, involving complex reasoning about the significance of the objects or scenes depicted. Typical query example is 'find pictures of a beautiful lady.'
The majority of content-based image retrieval systems mostly offer level 1 retrieval, a few experimental systems level 2, but none level 3.
Commonly used image features for content-based image retrieval were followings: 1) Color: color is one of the visual cues often used for content description, but most medical images are grayscale. Thus, color features are not used for medical image retrieval; 2) Texture: texture features mean spatial organization of pixel values of an image and used in standard transform domain analysis by tools such as Fourier transform, wavelets, Gabor or Stockwell filters. In the medical images, texture features are useful because they can reflect the details within an image structure; 3) Shape: shape feature has broad range of visual cues such as contour, curve, surfaces, and so on. Recently, many methods measures similarity between images using shape features has been developed.
Formerly developed commercial content-based image retrieval systems characterized images by global features such as color histogram, texture values and shape parameters, however, for medical images, the systems using global image features failed to capture the relevant information [9]. In medical images, the clinically useful information is mostly highly localized in small areas of the images, that is, the ratio of pathology bearing pixels to the rest of the images is small. Thus, the global image features such as color, texture, shape, etc. cannot effectively characterize the content of the medical images.
Initially, medical images were included in the content-based retrieval systems as a subdomain for trials [10,11]. Recent trials for content-based medical image retrieval were ASSERT system [12] for high resolution computed tomography (CT) images of the lung and image retrieval for medical applications (IRMA) system [13] for the classification of images into anatomical areas, modalities and viewpoints. Flexible image retrieval engine (FIRE) system handles different kinds of medical data as well as non-medical data like photographic databases [14].
In ASSERT system, the system lets the user extract pathology-bearing regions in lung images and these regions are then characterized by their grayscale, texture, and shape attributes, which are used for image retrieval.
The IRMA system (Figure 3) is a generic web-based x-ray retrieval system. It allows the user to extract images from a database given an x-ray image query. Local features and similarity measures are used to compute the nearest images. This system also developed a classification code for medical images to classify medical images based on four axes (modality, body orientations, body region, and biological system) [15]. The local features are derived from the previously classified and registered images that have been segmented automatically. IRMA analyzes content of medical images using a six-layer information model: raw data, registered data, feature, scheme, object, and knowledge. IRMA lacks the ability for finding particular pathology that may be localized in particular regions within the image.
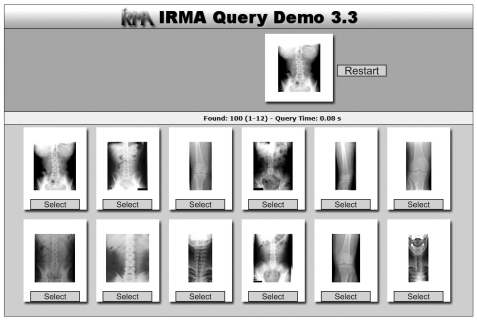
Image retrieval for medical applications (IRMA) content-based image retrieval results (http://irma-project.org/).
In FIRE system (Figure 4), query by example image is implemented using a large variety of different image features that can be combined and weighted individually and relevance feedback can be used to refine the result [16].

Flexible image retrieval engine (FIRE) content-based image retrieval results (http://thomas.deselaers.de/fire/).
The integration of content-based image retrieval system into the PACS system has been proposed [17,18]. However, an effective and precise medical image retrieval still remains a problem and recent researches aim at developing techniques that overcome this point.
IV. Controlled Vocabulary-Based System for Medical Image Retrieval
Currently, work is underway to create tools to enable semantic annotation of images using ontologies, providing an opportunity to enhance content-based image retrieval systems with richer descriptions of images. The suggested ontology systems are controlled terminologies such as Systematized Nomenclature of Medicine - Clinical Terms (SNOMED-CT) [19], Foundational Model of Anatomy (FMA) [20] and Radiology Lexicon (RadLex) [21].
1. SNOMED-CT Terminology
SNOMED-CT aims to be a comprehensive terminology that provides clinical content and expressivity for clinical documentation and reporting [19]. The SNOMED hierarchy is easy to compute, which was the primary reason for selecting the terminology for the research. SNOMED-CT has approximately 370,000 concepts and 1.5 million triples i.e., relationships of one concept with another in the terminology (Figure 5).
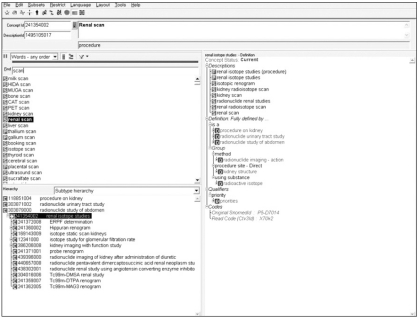
CliniClue Systematized Nomenclature of Medicine - Clinical Terms (SNOMED-CT) web browser (http://www.cliniclue.com/).
2. FMA
FMA is a reference ontology for the terms of anatomy and developed and maintained by the University of Washington and the US National Library of Medicine [20]. It contains 71,202 anatomical entities and more than 1,500,000 relations which form the machine processable standard vocabulary. FMA also provides definitions for conceptual attributes, part-whole, location, and other spatial associations of anatomical entities (Figure 6).
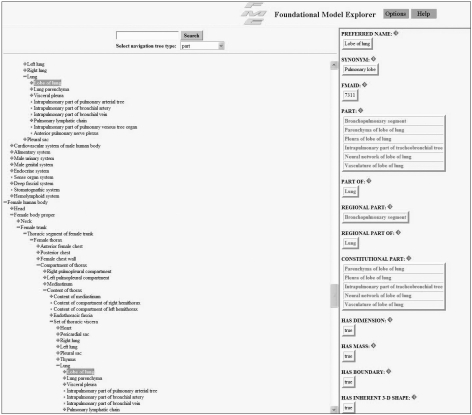
Foundational model explorer browser (http://fme.biostr.washington.edu/FME/index.html/).
3. RadLex Terminology
RadLex was developed to create a unifying source for medical imaging terminology by the Radiological Society of North America [22] and currently contains more than 32,000 standardized terms used in radiology reports (Figure 7). It contains not only domain knowledge but also lexical information such as synonymy. RadLex terminology helps the analysis of radiological information, allows uniform indexing of image databases, and enables structuring medical image information [23,24].

Radiology Lexicon (RadLex) term browser (http://www.radlex.org/).
V. Combined Text and Content-Based Medical Image Retrieval
Considering the intrinsic difference between the text and image in representing and expressing information, there have been approaches to combine the text-based and content-based image retrieval. Techniques that perform the text-based method first [25,26] and two methods at the same time [27,28] were studied.
The hybrid image retrieval systems to incorporate external knowledge that is encoded in lexicons, thesauri and ontologies were suggested [29].
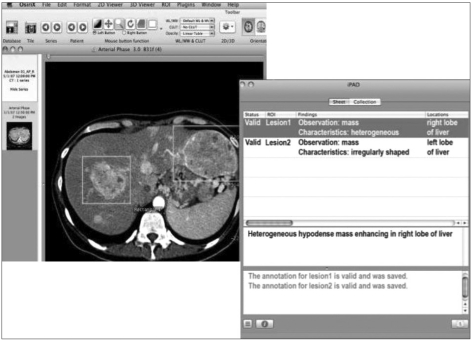
The new project, Annotation and Image Markup (AIM) for medical image annotation and markup is being developed to make all the key semantic (Figure 8) content of images machine-readable using controlled terminologies (mainly RadLex) and image markup standards [30].
VI. Conclusion
In the domain of medical imaging informatics, a huge amount of image data is being produced. A lot of work has already been done to improve the image retrieval systems. One is text-based approach and the other is content-based. Each method has its own advantages and disadvantages. Text-based method is widely used and fast, but it requires precise annotation. Content-based approach provides semantic retrieval, but effective and precise techniques still remains elusive.
Recently, a new controlled vocabulary, RadLex was developed to provide standardized terms for images and combined text and content-based methods were developed. For improved semantic image retrieval, it is proposed that image retrieval techniques be effectively integrated with external knowledge, annotation tools, and image markup systems.
In the near future, it is expected that the semantic contents of medical images will be totally computationally-accessible and reusable by the application of ontology and the development of new convenient tools.
Notes
No potential conflict of interest relevant to this article was reported.