Application of Social Network Analysis to Health Care Sectors
Article information
Abstract
Objectives
This study aimed to examine the feasibility of social network analysis as a valuable research tool for indicating a change in research topics in health care and medicine.
Methods
Papers used in the analysis were collected from the PubMed database at the National Library of Medicine. After limiting the search to papers affiliated with the National Institutes of Health, 27,125 papers were selected for the analysis. From these papers, the top 100 non-duplicate and most studied Medical Subject Heading terms were extracted. NetMiner V.3 was used for analysis. Weighted degree centrality was applied to the analysis to compare the trends in the change of research topics. Changes in the core keywords were observed for the entire group and in three-year intervals.
Results
The core keyword with the highest centrality value was "Risk Factor," followed by "Molecular Sequence Data," "Neoplasms," "Signal Transduction," "Brain," and "Amino Acid Sequence." Core keywords varied between time intervals, changing from "Molecular Sequence Data" to "Risk Factors" over time. "Risk Factors" was added as a new keyword and its social network was expanded. The slope of the keywords also varied over time: "Molecular Sequence Data," with a high centrality value, had a decreasing slope at certain intervals, whereas "SNP," with a low centrality value, had an increasing slope at certain intervals.
Conclusions
The social network analysis method is useful for tracking changes in research topics over time. Further research should be conducted to confirm the usefulness of this method in health care and medicine.
I. Introduction
The number of Science Citation Index (SCI) papers in the science and technology fields has increased 2.6-fold from 430,000 in 1974 to 112,000,000 in 2004, whereas that of SCI papers in health care and medicine fields indexed in the PubMed database increased 50% from 224,000,000 in 1999 to 380,000,000 in 2002 [1]. This number increased 13.6% in 2002 alone [2,3]. For example, a search of the keywords [tamoxifen AND breast cancer] in PubMed showed 6,750 results in seconds [3]. Papers published in Korea in the health care and medicine fields showed a two-fold increase from 64,000 in the 1980s to 113,000 in 2000 [4]. Therefore, it is not easy to obtain data from published papers and/or discern the correlations between the topics studied. Furthermore, it has become difficult for policymakers to understand the science behind research findings [5,6]. More recently, studies in the field have attempted to understand the intellectual structure by analyzing the social network by keywords. Researchers have analyzed the social network among different research topics such as preventative medicine [7] and epidemiology [8]. Researchers have also used specific field-related keywords as a unit of analysis to examine the change in research topics over time. A few studies examined the social network within the school of medicine in Korea [9] by extracting keywords from studies in different subfields such as medical information [10] and nursing [11]. Social network research in the field, conducted in the nation, has historically used researcher-extracted keywords or used indices for the analysis. Examples using social network analyses mainly used Medical Subject Heading (MeSH) terms by reviewing studies on influenza [12] and colorectal cancer [13]. The studies comprised panel research that employed MeSH terms to examine the changes in research topics over time.
To gain an understanding of the research context, researchers in various fields have used bibliometric analysis to conduct a co-citation analysis [14-16] and a co-word analysis [5,17-19]. The co-citation analysis has been applied in various fields to elucidate the intellectual structure in which interdisciplinary science knowledge is shared between different fields; namely, it was used to analyze the rate at which numbers of citations reduced (half-life) [7,20]. However, from this co-citation analysis, it is difficult to visualize the overall research picture and understand the detailed knowledge structure [7]. However, the co-word analysis permits retention of the information contained within the data and enables researchers to visualize it in simplified forms. Thus, this co-word analysis, based on the frequency of simultaneously appearing keywords, can be used to indicate linkages between research topics and offset the shortcomings of the co-citation analysis [17,21]. The co-word network analysis helps visualize the co-word analysis in a novel manner through a social network approach. The keywords are nodes, and simultaneously appearing keywords reflect the semantic relationships and research strategies. Thus, co-word network analysis helps users visualize concepts related to the topics, which enables researchers to conduct a content analysis [22]. This approach is a powerful tool for analyzing the knowledge structure of a 20-year-old database [5].
Co-word analysis has been used to measure the association strength between keywords to show research trends and patterns in polymer chemistry [17], nervous systems [23,24], software engineering [25,26], info search [21], and bioengineering [27,28]. Moreover, few studies have used this approach in the field of health care and medicine.
From 12 journals in medical informatics that were published from 1964 to 2004, Synnestvedt et al. [2] visualized the results of co-word analysis of MeSH terms from the PubMed database and co-citation analysis from the Web of Science index using Cite Space II. Research using co-word analysis has reported that "method" was an actively studied keyword. Moreover, the Web of Science index has reported that "practice guideline" and "patient safety" were the main research topics. One study analyzed 1,785 papers from the SCI for a co-citation analysis and identified the most frequently cited authors from the papers [29]. In the study, 1,506 papers from the PubMed database were analyzed to extract MeSH terms for the co-word analysis and to indicate that the John Cunningham Virus was a significantly influential factor in cancer. These studies were attempts to understand the differences in results obtained from the 2 different methods [2,29]. The co-word analysis in health care and medicine mainly consists of studies using MeSH terms from the PubMed database, which refer to medical terms defined by the National Library of Medicine (NLM) and are standardized by a closed circle of medical experts [28,29]. These studies aimed to identify leading research (research front) or emerging main keywords during a particular time-period. MeSH terms are similar in concept to bibliographic database keywords [29-31]. The network analysis of MeSH terms excludes unstandardized keywords and prevents unwanted indexer effects. However, extracting keywords included in the MeSH index become important for research. Moreover, keywords used by authors will have an indexer effect [32-34]. Furthermore, studies that use different keywords that have the same meaning must use an index extractor to standardize the keywords [21]. Therefore, for studies conducted by Korean researchers, knowledge maps were drawn up from extracted abstract keywords and classified as a thematic cluster [10,11,33]. Studies by Korean researchers have examined core word networks for preventative medicine [7] or epidemiology [8]. A study in nursing also used keywords extracted from 8 academic journals from 1995 to 2009 to examine research topic networks [11]. Using social network analysis, medical informatics study keywords were extracted from 1,075 research papers and proceedings published in 1995-2008 [10]. For these studies, researchers extracted keywords from papers or proceedings and used the index extractor to standardize the terms. Few studies conducted by Korean researchers used MeSH terms to extract keywords. Only recent research has used MeSH terms in social network analysis [12,13]. Thus, although the nature of the diseases varied, these studies found that research topics followed similar patterns or trajectories over time. Finding the disease causes was followed by sequence analysis, accumulation of sequence information, and efforts to manage and prevent diseases. Therefore, the present study aimed to apply social network analysis to the health care and medicine fields to examine changes in research topics over time.
Social Network Analysis
A social network is defined as a social structure comprising a set of actors (such as individuals or organizations) or networks of people related to one another (such as relationships, connections, or interactions) by particular characteristics [35]. Social network analysis is an interdisciplinary academic method that was developed by social psychologists and sociologists in the 1960s and 1970s. With the development of sophisticated and systematic analytical methods in computing and statistics, methods of social networks are currently widely used in economics, marketing, and industrial engineering [36,37].
Social network analysis identifies influential nodes, local and global structures, and network dynamics; namely, it transforms social networks into mathematical models of nodes and links from various data, proving, expressing, analyzing, and thus visualizing them or running simulations [38]. Social networks are visualized as graphs, and the actors' relationships are expressed with nodes and links. Nodes generally represent the actors, whereas links show transactions or exchanges between 2 actors in the network. The core concepts in social network analysis are degree and density. Degree refers to the number of connections a node has in a network, and individuals with many connections can mobilize a large amount of resources and play a central role in the flow of information [39,40]. Density refers to the ratio of the number of actual connections to all possible connections. Density has an inverse relationship to group size. Therefore, a person with same number of connections may see his density decrease as group size increases. It is necessary to standardize group size to compare the density of people in groups of different sizes [39,41].
The use of social network analysis is not limited to people; rather, it can have variable properties. Keyword research can be considered a variation of its properties. If a node is made to represent a keyword, then a keyword with high-degree connections becomes an actively researched topic in the field. Keyword correlations may be differentiated by degrees. This may be interpreted as influence, which is measured by the centrality of keywords in the network. Centrality can be measured by counting the number of connections a node has or the number of steps one needs to take to reach every node [39,42]. Therefore, a research topic related to many others in the network or one with few steps to reach one's topic could be said to have centrality. Hence, the indicator for centrality was proposed [43]. Freeman proposed 2 kinds of centrality: local centrality, which is high if a node has many direct connections; and global centrality, which is measured by a node's strategic position within a correlational context [43]. A node with high local centrality may also be one with high global centrality, but the 2 do not have to be identical [35,41,43]. For example, "Adenocarcinoma" and "Influenza Human" have high degrees of local centrality in their respective fields of colorectal cancer and influenza. "Risk Factor," on the other hand, may not be an important research topic with high local centrality in either of those fields, but it probably has high global centrality. Degree is a good indicator of local centrality. However, it is not easy to compare degrees across different groups because degree is represented as a percentage of total connections in the network. Therefore, degree is usually compared between same-size groups, or additional steps are required to standardize size [39,41]. In the present study, the authors used local centrality to compose a network of core keywords and standardized degrees of local centrality.
II. Methods
1. Data Extraction and Keyword Selection
On September 3, 2011, the authors collected data from the PubMed database of the NLM and limited papers affiliated with the National Institutes of Health (NIH). A total of 27,125 papers published in 1967-2011 were selected for the analysis. From these papers, excluding subheadings of the keywords indexed in MeSH, a total of 256,613 keywords were extracted. Duplicates and "check tags" were also removed, leaving a total of 13,424 keywords. To analyze core keywords, the most commonly used keywords ranked in the top 100 list were selected first. The authors then checked whether any keywords were omitted from the list by sorting the selection into six 3-year intervals to examine keyword trends over time. The authors grouped papers up to 1995 into 1 group because of the low number and observed that the keywords remained consistent over time. A keyword expert was then consulted to confirm this observation. After duplicate terms were removed, a total of 748 keywords from the top 100 list were left, and 190 keywords were selected for the study [13].
2. Network Analysis for Research Topics
To compose the social network of selected keywords, this study used NetMiner V.3. A network with weighted degree centrality was constructed [44]. Keywords are expressed by nodes. For keywords that appeared on more than one paper focusing on different subjects, a matrix was drawn to assume linkages between research subjects. For example, a 190 × 190 keyword matrix was drawn for NIH. To examine the change in core keywords, degree centrality for the analysis was used. Degree centrality is used to construct a keyword centrality index to indicate a node's centrality in the network. It adds the number of direct connections a node has based on the number of nodes in the network [43,45]. As the number of links between the nodes increases, the index grows larger. Weighted degree centrality adds centrality to those nodes with more direct connections. This study uses weighed degree centrality to analyze core keywords. Keywords with higher degree centrality are more actively researched keywords in the field [35,46].
1) Keyword network analysis
To observe changes in keywords over time, the keywords were divided into 3 different time intervals and social networks were constructed for each time interval. The authors categorized keywords that appeared before the year 2000 into one group and divided the other years into 3-year intervals. The last group of keywords was categorized into 2009-2010. The authors used the pruning method, which creates a social network using core keywords with high degrees of connection, to observe any change in the core keywords between intervals. A cut-off is used to recreate a social network with only those core keywords with high connection levels [6,16]. For example, "pruning at 100" discards those keywords with values <100 and recreates the network using the remaining keywords. Information is lost in the process, but researchers can observe the relationship between core keywords with strong degrees of centrality [35]. The cut-off was set at 0.6% of the highest degrees. A previous study has used 0.1% [13], but the authors could not use the same cut-off because the number of core keywords left thereafter would be too small. At 0.6%, the authors could observe the relationship between 40-50 nodes [13] and could observe the emergence and decline of keywords between the time intervals and the changes in research topics over time [16].
2) Change in keyword slope
To compare differences between the set intervals, 20 keywords from each time interval with the highest degree of centrality were extracted. Duplicates were removed, and the remaining 35 core keywords were observed to examine the changes between the intervals. For comparison between the intervals, the authors adjusted the values of the degrees of centrality to consider group size differences [35]. Therefore, the following formula to calculate standardized degrees of centrality for the time intervals was noted:
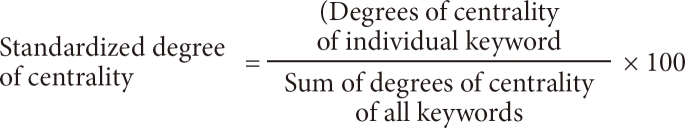
NCVi = ICVi / (ΣCVi) × 100
(NCVi = nomalized centrality value, ICVi = individual centrality value, ΣCVi = total centrality value).
For example, the centrality value for the keyword "Risk Factor" was 5.24 and the sum of the centrality value was 370.04. Therefore, the normalized centrality value equals:
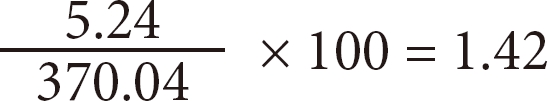
To examine standardized degree centrality values to measure the change in core keywords, the authors used statistical package SAS 9.1.3 (SAS Inc., Cary, NC, USA), and regression analysis of the centrality value was used to observe changes in the slope of each keyword (Table 1).
The regression function is expressed the following way:
Yi = αi + βiX + εi
(α = constant, β = slope, ε = error; 0 for this function).
The equation used for the regression analysis was as follows:
Yi = αi + βiX
(Y = value of slope for individual keyword, X = year).
III. Results
1. Network Analysis for Research Topics
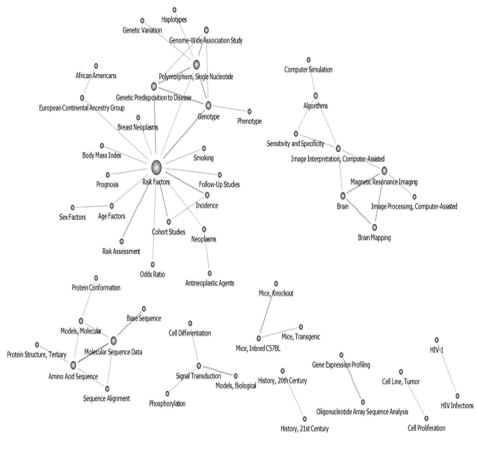
Of the 190 core keywords selected, the one with the highest centrality values was "Risk Factors," followed by "Molecular Sequence Data," "Neoplasms," "Signal Transduction," "Brain," and "Amino Acid Sequence." The authors used the pruning technique on the 190 keywords, and the cut-off value was set at 100. "Risk Factors" was found to have the highest centrality value in the entire network. The following 3 networks were also formed: a "Molecular Sequence Data"-related network, a "Gene Expression Profiling"-related network, and a "Brain"-related network (Figure 1).
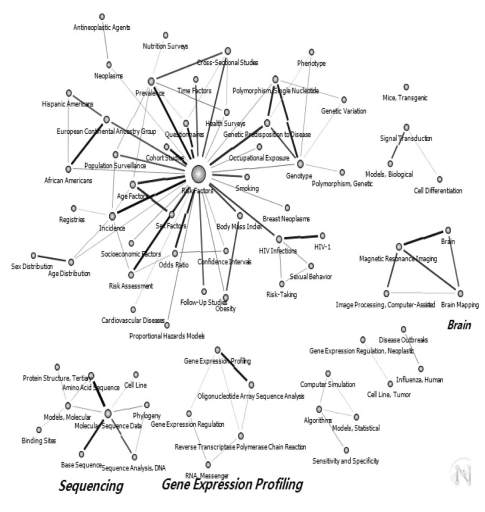
Keywords network of 190 in National Institutes of Health after pruning off 100 degree below (1967-2010, n = 61).
1) Keywords network analysis according to time intervals
Papers published in different time periods were used to observe how research topics changed over time. Topics were divided into different time intervals: those published before the year of 2000 and those in 3-year intervals thereafter. The authors observed each time period and also made adjustments to compare groups of different sizes. The maximum centrality value was set at 0.6%, and the top 20 keywords from each time intervals were selected (Table 2).
Topics before the year 2000 were pruned at 25; therefore, 57 keywords were obtained, and these were used to recreate the network. Keywords such as "Molecular Sequence Data," "Amino Acid Sequence," "Base Sequence," "RNA, Messenger," and "Tumor Cells, Cultured" had high centrality values, and all of the keywords were connected to form one large network. Researchers used sequencing analysis data to conduct research in "Apoptosis" and "Signal Transduction" in search of "Anti Neoplastic Agent." "Brain" and "HIV Infection", however, were separated in the network (Figure 2).
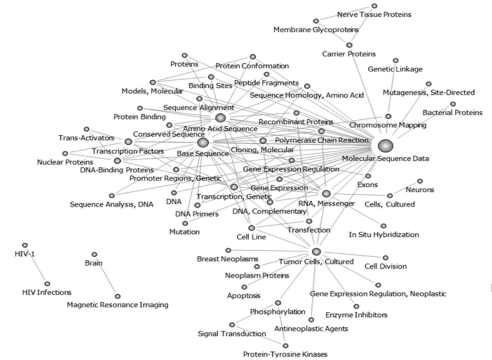
Keywords network in National Institutes of Health After pruning off 25 degree below (before 2000 year, n = 57).
Topics in the 2000-2002 time interval were pruned at 28, and 60 keywords were obtained. Cancer grew as a research area during this period based on "Sequencing" and "Tumor Cell". Research on "Brain" was expanded, and "Risk Factors" emerged as a new area of study (Figure 3).
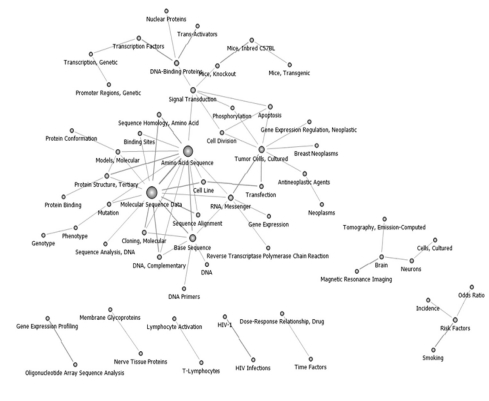
Keywords network in National Institutes of Health after pruning off 28 degree below (2000-2002, n = 60).
Networks were divided in the 2003-2005 time interval. Social networks around "Sequence Analysis" decreased, whereas the "Gene expression profiling" network started to grow. "Risk Factor" was connected to different areas of interest such as "Breast Neoplasm" and "Genetic Predisposition to Disease." "Tumor Marker" was newly linked to "Anti Neoplastic Agent." These changes confirmed that cancer research began moving in the direction of chemotherapy, diagnostics, and prediction (Figure 4).
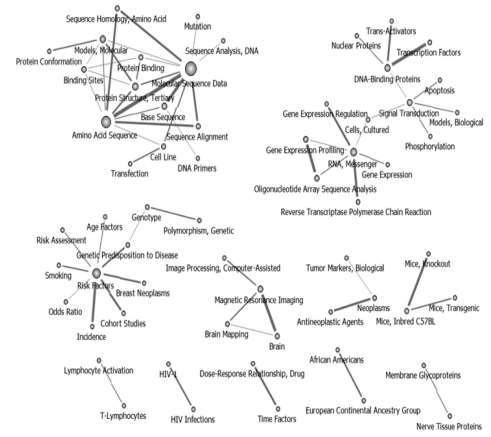
Keywords network in National Institutes of Health after pruning off 30 degree below (2003-2005, n = 60).
The authors selected 60 nodes from the 2006-2008 interval by pruning at 27. During this time period, the networks formed in this period showed a clear difference from those found in other periods. The keywords did show strong linkages or centralities. As a result, the networks were divided into hubs. Research in "Risk Factor" grew with increasing interest in genetic predisposition. The introduction of "SNP" may indicate that research interest in sequencing further declined while interest in "Brain research" grew during this period (Figure 5).
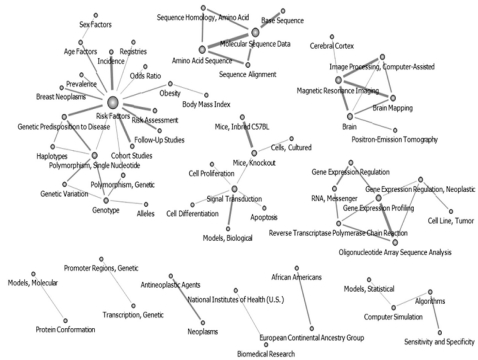
Keywords network in National Institutes of Health after pruning off 27 degree below (2006-2008, n = 60).
A total of 53 nodes were selected to construct a social network for the 2009-2010 period. Pruning was performed at 27. "Risk Factor" research expanded indirectly through growth in "Genetic Predisposition to Disease" and "SNP." The centrality of SNP grew with the publication of the genome wide association study (GWAS). Moreover, "Anti Neoplastic Agent" was absorbed into the "Brain"-related network (Figure 6).
2) Comparisons of centrality and slope
The authors divided the core keywords into 3-year intervals and selected the top 20 keywords with high degrees of centrality from each interval. The duplicates were removed. The authors used these keywords to compare the observed changes in research topics. After accounting for differences in size, statistical package SAS 9.1.3 (SAS Inc.) was used to conduct the research on observed changes in degree and centrality.
"Risk Factors," "Genotype," "Genetic Predisposition to Disease," "SNP," and "Cell Line, Tumor" showed positive increases in both degree and centrality. "Molecular Sequence Data" and sequencing had rapidly decreasing negative values over time (Table 1).
Although "Molecular Sequence Data" listed as a top keyword showed a rapidly decreasing slope, it was not statistically significant (p = 0.1403). Therefore, the changes between intervals should always be checked, even for the top 20 keywords with high centrality values. As recent as 2005, "SNP" was not one of the top 20 keywords; however, its centrality and slope increased rapidly in the 2006-2008 time interval. "SNP" showed a rapidly increasing slope that was statistically significant (p < 0.05). The authors checked all the keywords with an upward slope and were able to confirm that "Genotype" (p < 0.05) was related to "Risk Factor" and "Magnetic Resonance Imaging" (p < 0.01), all of which had rapidly increasing slopes.
IV. Discussion
Social network analysis is widely used in various disciplines. This study extracted keywords from NIH papers to conduct co-word analysis. Changes in research topics can be identified effectively through social network analysis.
This research used the PubMed database of the NLM. Studies on network analysis typically used the SCI or Scopus database to measure the influence of academic journals using indices drawn from science databases. Limiting the research scope to a number of influential journals can increase the reliability of the research outcome. Because these databases offer citation subject classification services, researchers use co-citation analysis to understand research trends within a given subject using social network analysis with cogitation analysis; thus, one can easily visualize research trends in a field [47,48]. However, health and medicine researchers have relied on the PubMed database to conduct co-word analyses of detailed subjects [2,13]. Papers listed in the PubMed database are reviewed by Medline and are then given MeSH-indexed terms [49], which are similar in concept to a bibliographic database. MeSH indexes and ensures consistency in medical papers [30,31,50]. Therefore, the MeSH index is more consistent and systematic than other databases. MeSH terms are divided into headings, main headings, subheadings, geographic headings, check tags, methodology publication type, and other categories. Except for subheadings and main headings, however, most indices need to be standardized for classifications [30]. Moreover, the MeSH index quality varies considerably. Researchers may need to take steps to standardize the index considering various issues when extracting and standardizing terms. Unnecessary parts certainly need to be removed. From the start, check tags were removed. One may also need to consult experts when consolidating MeSH terms [51].
The authors analyzed a network of core keywords to understand research trends in the study field. The social network research using centrality measures are not typically weighted [44]. This study uses weighted measures for linked keywords as well as a number of connections to understand the centrality of research topics. Thus, the number of connections is accounted for in the weighted value [45,52] As a result, subject areas were illustrated in which active research is being conducted. However, it is unclear whether this measure will improve research outcomes. Many adjustments are required to use the weighted measure. Major topics may be useful for analyzing keywords of papers collected from PubMed. Using weighted values for major topics may be a good alternative [52]. Additional research is needed to validate the results of this study. The authors could not illustrate results of all keywords; therefore, they chose the ones with high frequency to create the networks and also used pruning to select those with high centrality. Therefore, the keywords currently shown in the network must be excluded. This is a limitation of social network analysis; as such, one must be careful in making the selection.
The social network analysis of 190 keywords divided into intervals showed that "Risk Analysis" was the most commonly researched topic in the health and medicine field over time. This finding illustrates that the NIH is sure of its role as a public health institution, and the use of keywords such as "Molecular Sequence Data," "Neoplasms," and "Signal Transduction" illustrates public administrators' responsibility as one who conducts basic research in the field. Moreover, the study shows that the major research in the field shifted from "Molecular Sequence Data" to "Risk Factors." The study also showed how research topics are being expanded into various areas, and thus, are adding new keywords. For example, in the network based on "Risk Factors," new keywords such as "GWAS" via "SNP" emerged. In addition, "Anti Neoplastic Agent" was indirectly absorbed into the main network via "Neoplasm"; therefore, increasing local centrality is the key to social network analysis. A node must increase its centrality to be more useful to those around it.
For this research, directly connected keywords were representative of well-researched areas. However, the study showed that the "Sequence Analysis" network was in decline. Therefore, this study showed that the keyword network could be explained in terms of the actor (node) and the degrees of relationship (degree). Moreover, research topics in the field of medicine followed a common pattern: a researcher was first interested in analyzing DNA of diseases (base sequence, amino acid sequence) and was then interested in collecting more information about the disease. With developments in medicine, researchers have become more interested in analyzing treatment outcomes. The period was followed by interest in improving diagnoses, predicting stages of development, and finding risk factors. Subsequently, researchers became much more interested in preventative measures. A similar pattern could be observed in the field of intestinal cancer, the development stages of which are relatively well known.
A centrality value alone does not guarantee that the keywords are actively researched. Checking for slopes helps verify the results. "Molecular Sequence Data" had high overall centrality; nevertheless, this study showed that its slope declined in an interval analysis. On the other hand, "SNP" had a generally low centrality value; however, its slope increased in an interval analysis. Thus, checking a keyword's slope can reduce such oversights. However, this study did not use clustering; instead, the authors focused on the correlations between individual keywords to construct the networks. This approach can lead to data loss. Losing data is an inherent problem of social network analysis research. One needs to be clear about the scope and definition used in the analysis and carefully interpret the results.
By analyzing the NIH papers, this study demonstrated that NIH's keyword with the highest centrality values was "Risk Factors" and research previously conducted was obviously related to public health. This study also showed that research on personalized treatments and the risk factors of diseases through genetic analysis has been actively conducted over time. In addition, the dynamic change in keywords was observed. As time passed, research on organisms actually has evolved and perished. In the literature, researchers have examined studies on influenza [12] and colorectal cancer [13], both of which used network analysis to identify a certain pattern of research topics over time. Therefore, the findings of this study suggest that the social network analysis can be applied to research in health care sectors. Applying this approach ultimately would enable us to propose a milestone for strategic planning of research in stages. It is also suggested that future research should incorporate experts review in the field to develop evidence-based research. Finally, a time series analysis of an individual disease should be used to define the developmental stages or turning points of the disease [16,29].
Acknowledgments
This research was supported by the grant no. 182 from the Korea Centers for Disease Control and Prevention. This research also was partially supported by the FY2010 intramural research fund from the Chungbuk National University.
Notes
No potential conflict of interest relevant to this article was reported.