Tuberculosis is an important global public health problem. Ranked above HIV/AIDS, it is considered the leading cause of death by a single infectious agent. According to the Global Tuberculosis Report 2020, tuberculosis causes 1.3 million deaths per year [
1]. Although the mortality and incidence rates of tuberculosis in Korea have steadily decreased in recent years [
2], the management of tuberculosis remains challenging and requires more effective efforts. In clinical settings, inadequate treatment leading to therapeutic failure and drug resistance was reported as a major challenge in tuberculosis management efforts [
3,
4]. Therefore, precision tuberculosis treatment strategies should be deployed to reach the global tuberculosis control targets.
The development of a centralized and cost-effective platform that can improve tuberculosis management is an urgent need. However, recent studies on tuberculosis-related digital technology have focused on diagnostic tools or treatment adherence technologies and have not expressed interest in developing systems that support conducting research and implementing high-precision, personalized treatment [
5]. In this article, we introduce a system named the ŌĆ£Center for Personalized Precision Medicine of Tuberculosis: Smart Research and Development Workstation (cPMTb Smart R&D Workstation).ŌĆØ The cPMTb Smart R&D Workstation, which is available at
https://smart.cpmtb.kr/, is expected to become a proof-of-concept for data-driven personalized precision tuberculosis treatment. The overall architecture and information technology of the system are illustrated in
Figure 1.
For data collection and generation, tuberculosis patients receiving treatment with anti-tuberculosis drugs were recruited from each hospital site. Subjects who were prisoners, could not provide voluntary commitment, or were pregnant were excluded. All recruited patients provided written informed consent. A whole blood, plasma, or dried blood spot (DBS) sample of each patient was collected and transferred to the cPMTb laboratory. From those samples, anti-tuberculosis drug concentrations were simultaneously measured using our previously established method [
6]. The drug concentrations measured from DBS were converted to plasma drug concentrations using our in-house method. These data play a critical role in therapeutic drug monitoring-based dose adjustment model development and personalized medicine [
7]. We also actively genotyped
NAT2 and
SLCO1B1 genes, as their variations have been reported to be associated with unfavorable outcomes in tuberculosis patients [
8,
9]. Additionally, minimum inhibitory concentration assays were conducted when
Mycobacterium tuberculosis isolates were available.
With higher expectations of achieving personalized medicine in tuberculosis treatment, a comprehensive and diverse type database is critically important for further precision model-informed dosing development, both for pharmacometrics and for machine learning approaches. Therefore, we developed a database that encompasses multiple data domains, types, and features, as illustrated in
Table 1. As of March 24, 2022, a total of 2,123 tuberculosis patients have been recruited from 21 hospital sites, including information on 22 anti-tuberculosis drugs (
Supplementary Table S1). Detailed information is presented in real-time at
https://smart.cpmtb.kr/#/cohort/status.
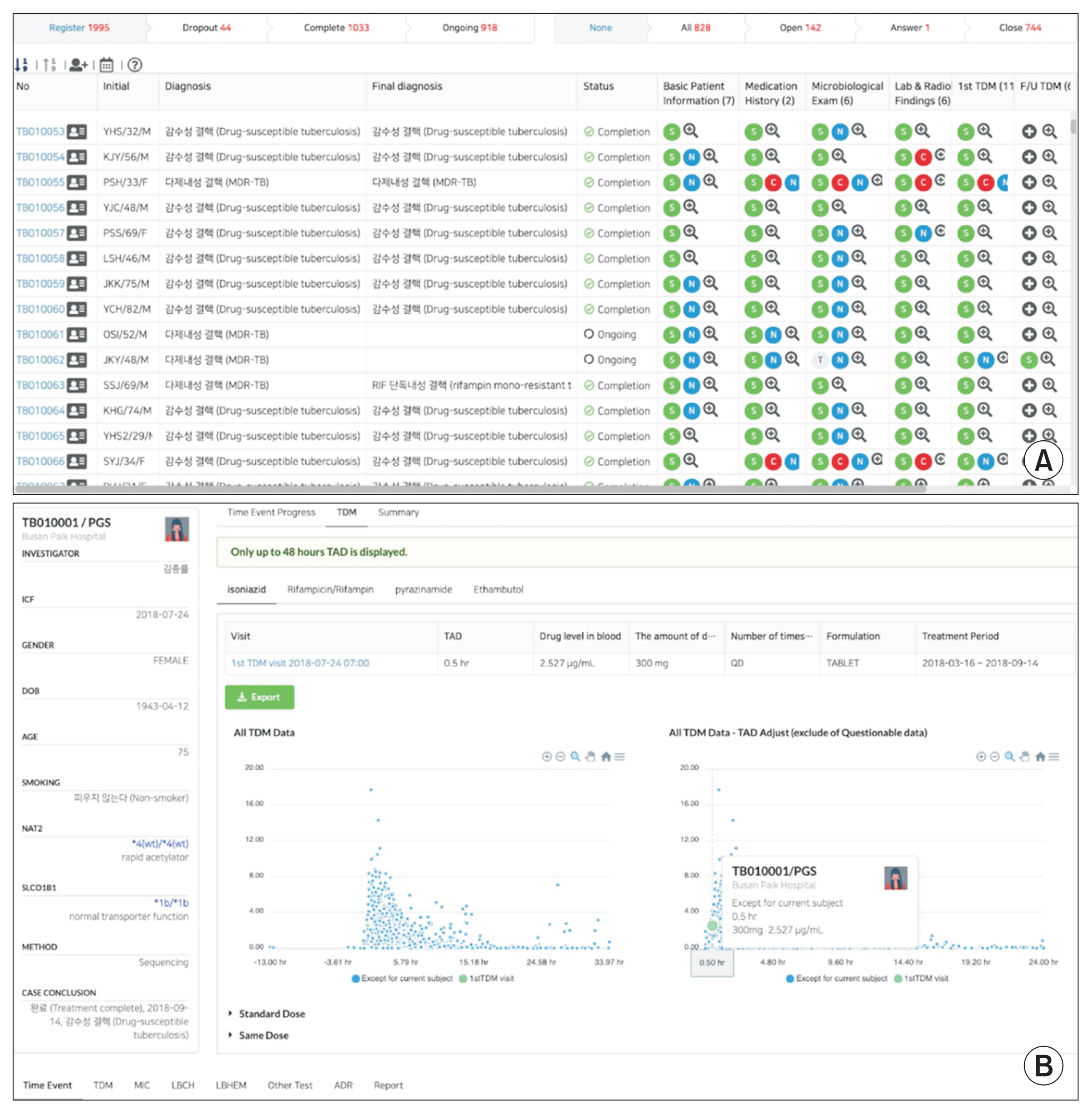
For data visualization, a comprehensive review including diagnosis, status of treatment, essential characteristics, medication history, microbiological examination results, laboratory and radiological findings, drug concentrations, adverse drug reaction information, and other monitoring data of each patient is provided. These parameters are useful for researchers to examine data from an individual-oriented perspective. We also developed a function to generate a drug concentration time profile, which provides information on the pharmacokinetic aspects of a given drug based on multiple conditions (
Figure 2). This has inspired investigators to initiate pharmacokinetics-related research projects [
10,
11].
From a broader perspective, a single country or single-center effort is not sufficient to achieve the World Health OrganizationŌĆÖs End Tuberculosis Strategy milestone. Instead, collaboration of multiple nations and multiple centers is required to reach the goal of personalized, high-precision tuberculosis treatment. We aim to expand our cohort to other hospital sites in Korea and worldwide. When local sites collaborate with or join our study, they receive support in measuring the concentration of anti-tuberculosis drugs and genotyping well-known variants related to anti-tuberculosis drug response. Importantly, sites can also participate in our research to develop model-informed precision dosing for tuberculosis treatment. This collaborative model saves substantial amounts of human labor and other resources.
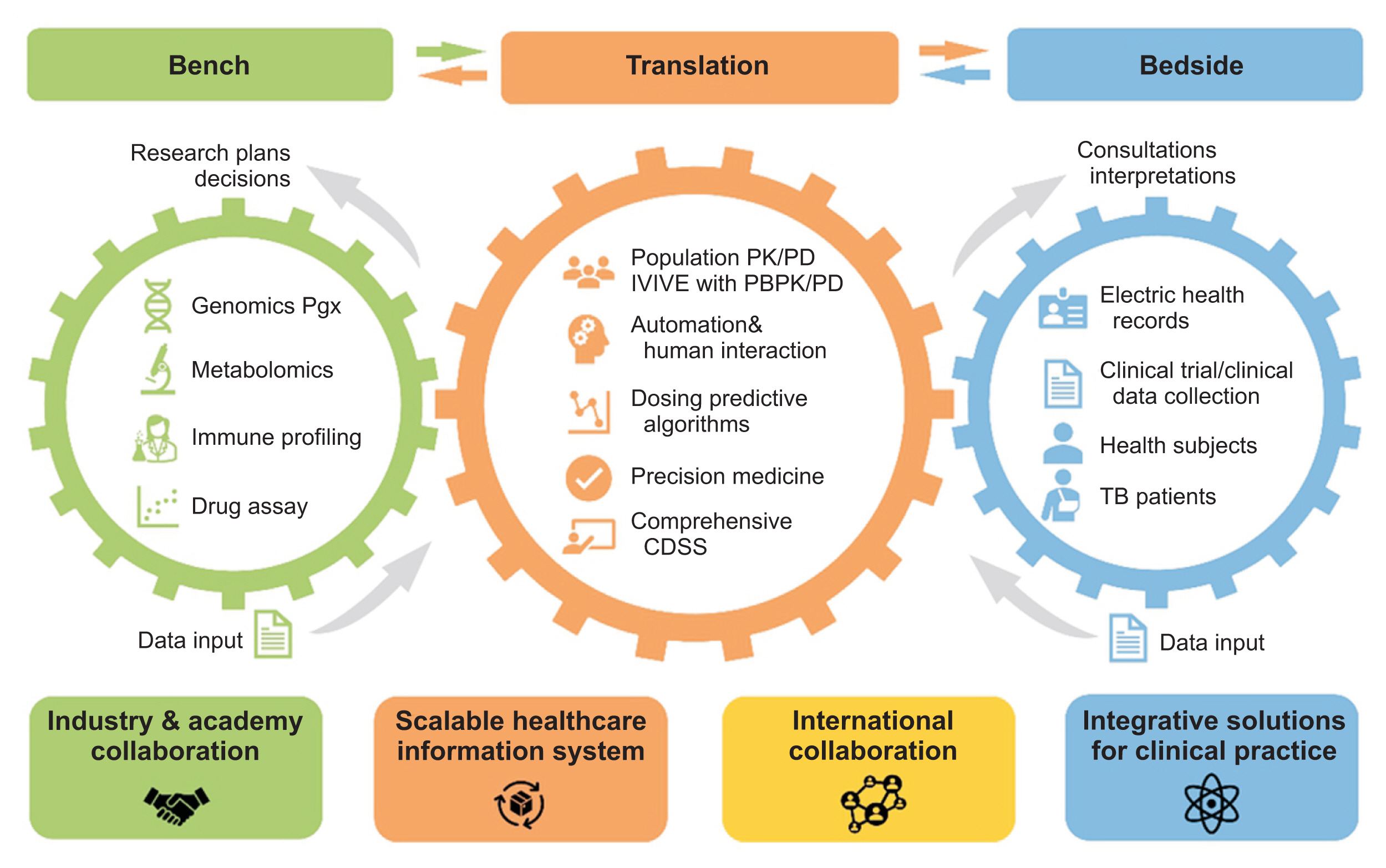
As an action plan, the genetic profile of all patients will be genotyped using a genome-wide screening array, including pharmacogenomics (PGx) and absorption, disposition, metabolism, and elimination (ADME) panels. Additionally, we regularly update the database with information related to ADME and PGx of all anti-tuberculosis drugs from the literature. Taking these considerations into account, many discovery and confirmatory studies can be conducted to identify potentially relevant biomarkers for individualized treatment. To summarize, we anticipate that our system will be an essential component of global efforts to transfer knowledge from the bench to the bedside and to actualize personalized precision tuberculosis management (
Figure 3).
In summary, we briefly demonstrated the cPMTb Smart R&D Workstation and its functionality. The system is expected to facilitate an approach to high-precision personalized treatment of tuberculosis through the integration of multiple data domains, seamless management, comprehensive data visualization, and model-informed dosing development. We also anticipate expanding our multinational collaborations, thereby strengthening ongoing efforts to accomplish the World Health OrganizationŌĆÖs End Tuberculosis Strategy.
Supplementary Materials
Table S1. Number of anti-tuberculosis drugs and number of patients being treated by each drug (retrieved on March 24, 2022)
Table S1. Name of hospital sites and IRB numbers
Table S1. List of participating investigators from each hospital site
hir-2022-28-2-176-suppl.pdf
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2018R1A5A2021242).
The authors would like to acknowledge all investigators participating in the project. The list of investigators is given in
Supplementary Table S3. Additionally, we also appreciate all staff who participated in the study.
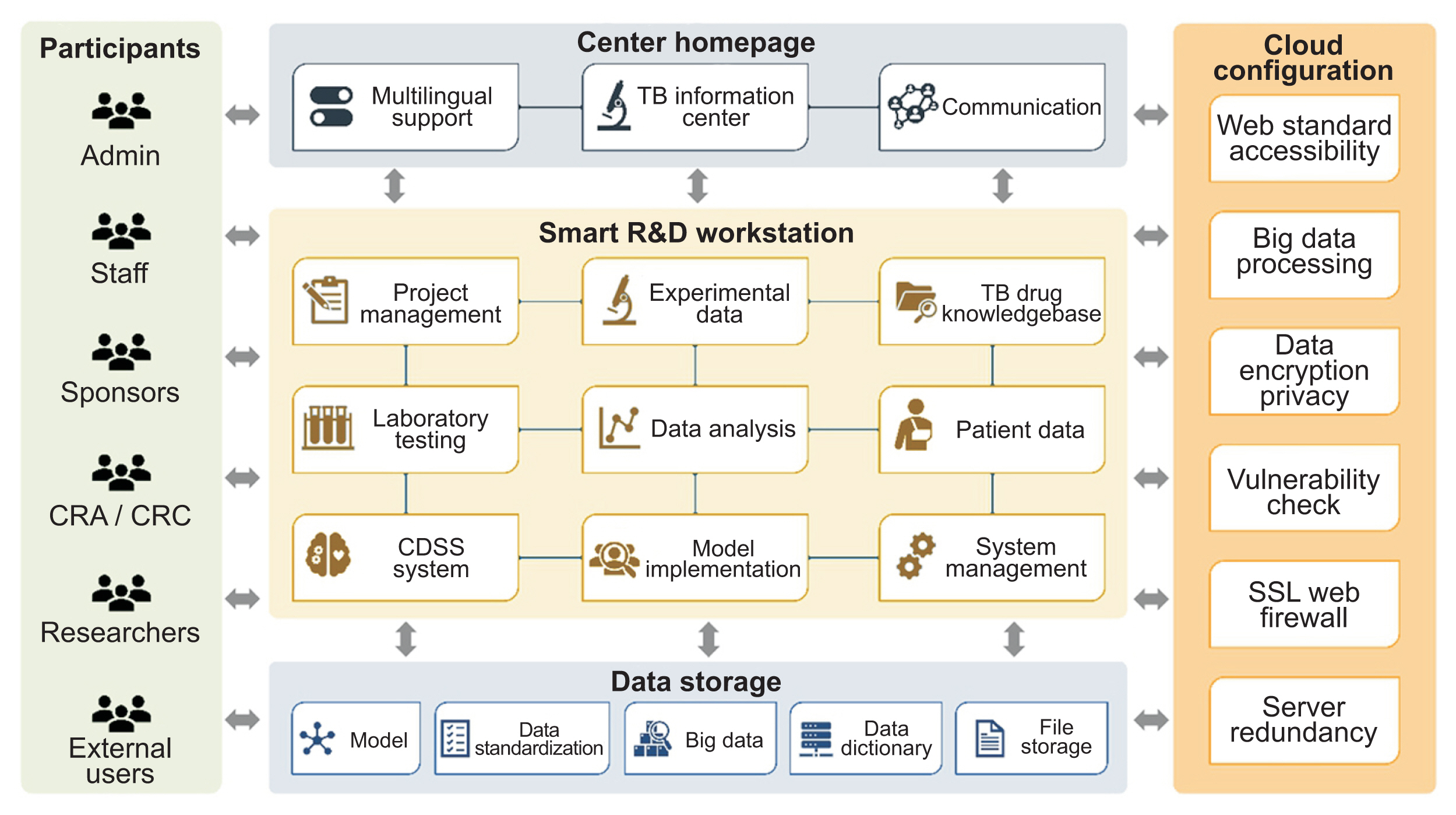
Figure┬Ā1
The cloud-based architecture of the cPMTb Smart R&D Workstation (Center for Personalized Precision Medicine of Tuberculosis: Smart Research and Development Workstation). CRA: clinical research associate, CRC: clinical research coordinator.
Figure┬Ā2
Overview of functions of the cPMTb Smart R&D Workstation (Center for Personalized Precision Medicine of Tuberculosis: Smart Research and Development Workstation). (A) Clinical case review screen for an individual patient. (B) Interactive scatter plots of drug concentrations with various options for filtering and normalization.
Figure┬Ā3
Bi-directional flow of information in the cPMTb Smart R&D Workstation (Center for Personalized Precision Medicine of Tuberculosis: Smart Research and Development Workstation) for personalized medicine of anti-tuberculosis drugs. CDSS: clinical decision support system, PK/PD: pharmacokinetics/pharmacodynamics, IVIVE: in vivoŌĆōin vitro extrapolation, PGx: pharmacogenomics, ADME: absorption, distribution, metabolism, and elimination, PBPK/PD: physiology-based pharmacokinetics/pharmacodynamics.
Table┬Ā1
Major data domains of the cPMTb cohort database
|
Domain |
Main content |
|
PatientsŌĆÖ information |
Basic characteristics (age, body weight, height, ŌĆ”), Co-morbidity, Co-medication, Smoking status, Ethnic, etc. |
|
Laboratory testing |
Biochemistry testing (ALT, AST, ALP, CR, eGFR, ŌĆ”), Hematology testing (WBC, RBC, PLT, HGB, HCT, ŌĆ”) |
|
Diagnosis |
Location, Drug susceptibility test, Xpert test, X-ray results, Relapse, Other tests |
|
Treatment |
Tuberculosis treatment history, Regimen, Duration, Change regimen |
|
Sample collection |
Time of collection, Type of sample, Biobank location, Status of sample |
|
TDM |
Time of TDM, Drug concentration, PK analysis, TDM report |
|
ADR |
ADR term (MedDRA: PT, MedDRA: SOC), Time of onset and offset, Severity, Suspected drugs, Action |
|
Multi-omics |
Genotype of NAT2, SLCO1B1, Genome wide screening array, Transcriptomics, Metabolomics
|
|
MIC |
Isolations, MIC assays, MIC distribution |
|
TB drug data |
in vitro data, in vivo data, ADME collected data, Available PK parameters |
|
Models |
Population pharmacokinetic models, PBPK models, AI/ML models |
|
Treatment |
outcome Time of treatment, Compliant or noncompliant, Recure or not |
References
2. Korea Disease Control and Prevention Agency. Annual report on the notified tuberculosis in Korea, 2019. Cheongju, Korea: Korea Disease Control and Prevention Agency; 2020.

3. Lange C, Dheda K, Chesov D, Mandalakas AM, Udwadia Z, Horsburgh CR Jr. Management of drug-resistant tuberculosis. Lancet 2019;394(10202):953-66.


4. Furin J, Cox H, Pai M. Tuberculosis. Lancet 2019;393 (10181):1642-56.


6. Kim HJ, Seo KA, Kim HM, Jeong ES, Ghim JL, Lee SH, et al. Simple and accurate quantitative analysis of 20 anti-tuberculosis drugs in human plasma using liquid chromatography-electrospray ionization-tandem mass spectrometry. J Pharm Biomed Anal 2015;102:9-16.


11. Soedarsono S, Jayanti RP, Mertaniasih NM, Kusmiati T, Permatasari A, Indrawanto DW, et al. Development of population pharmacokinetics model of isoniazid in Indonesian patients with tuberculosis. Int J Infect Dis 2022;117:8-14.














