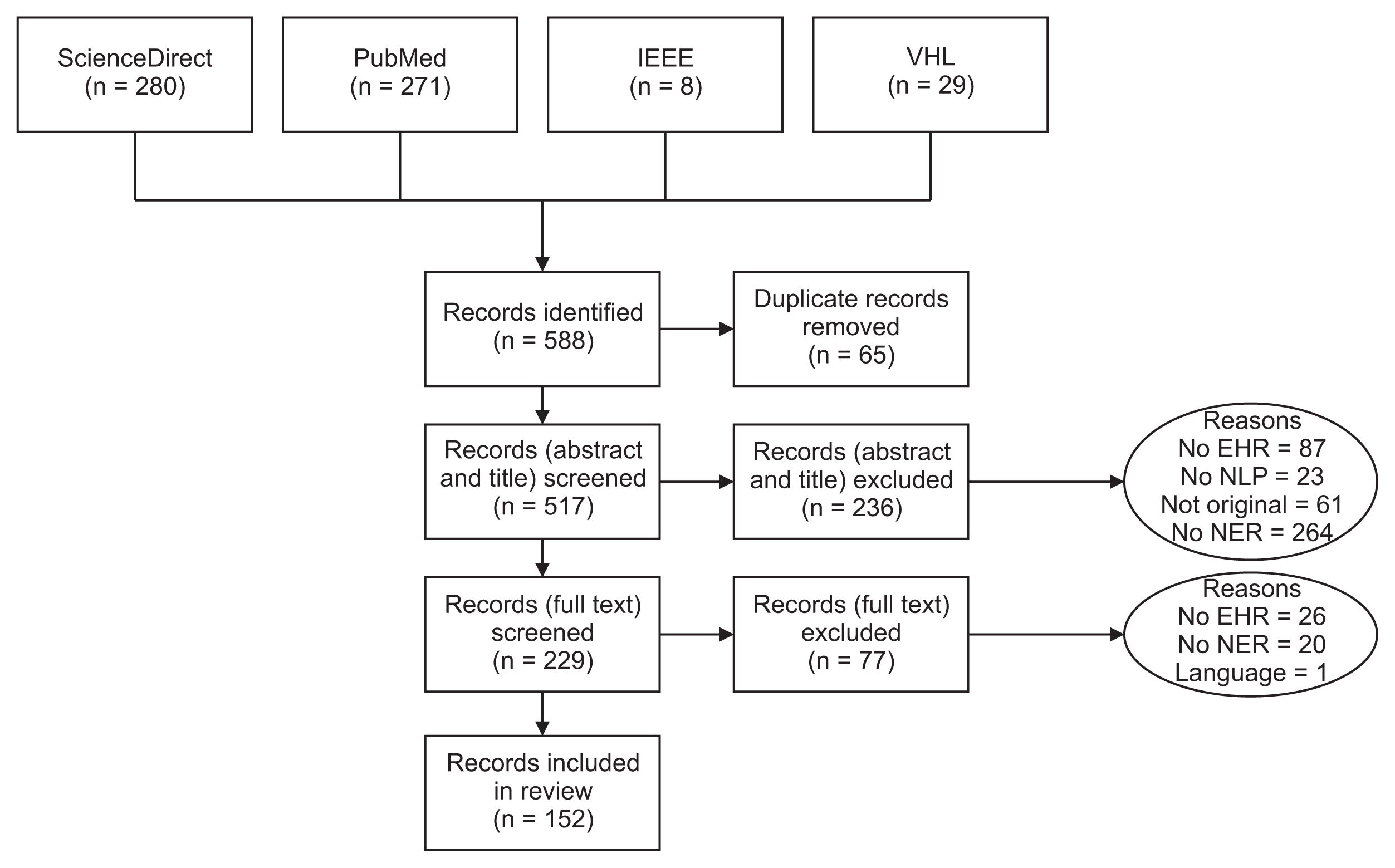
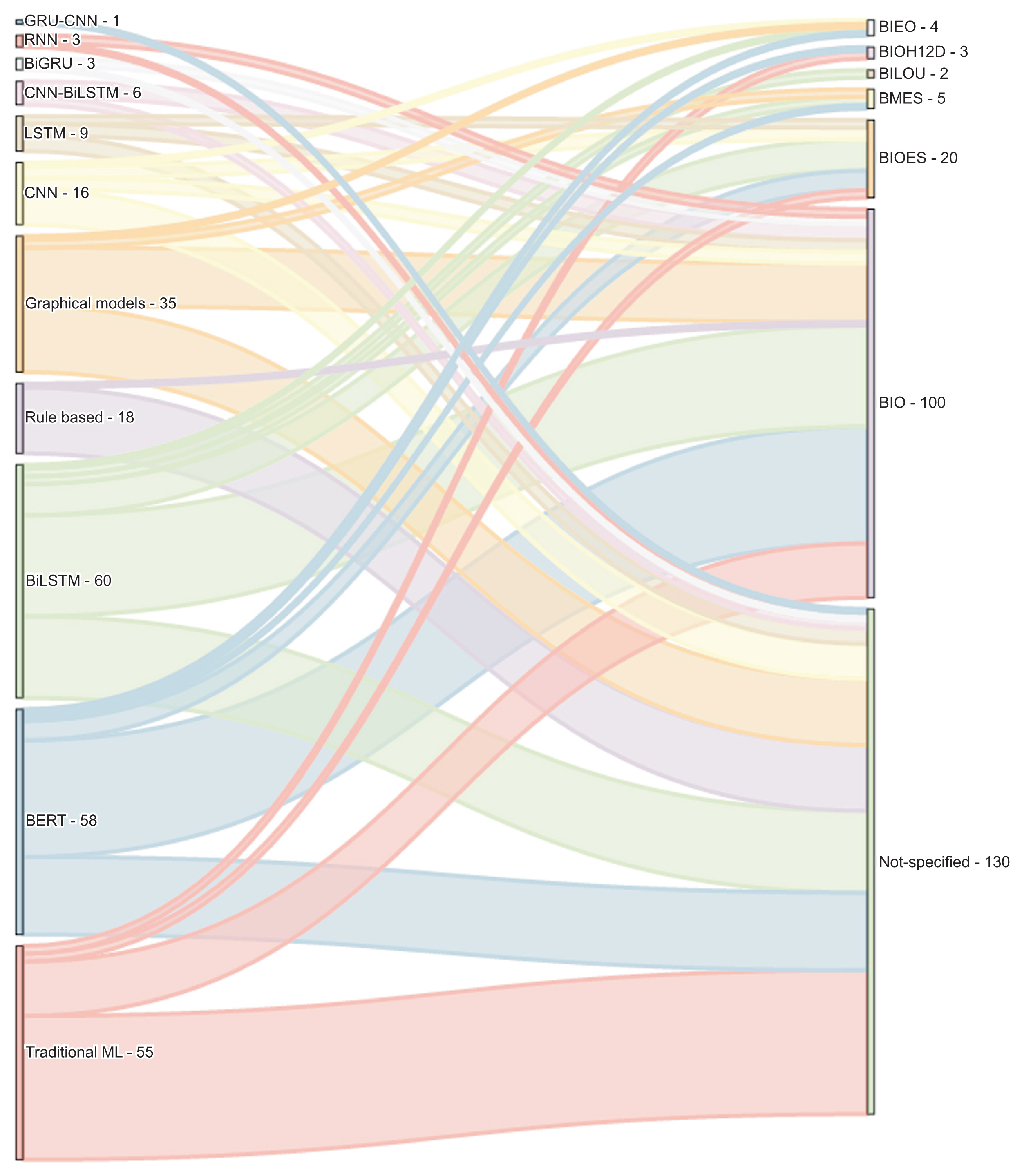
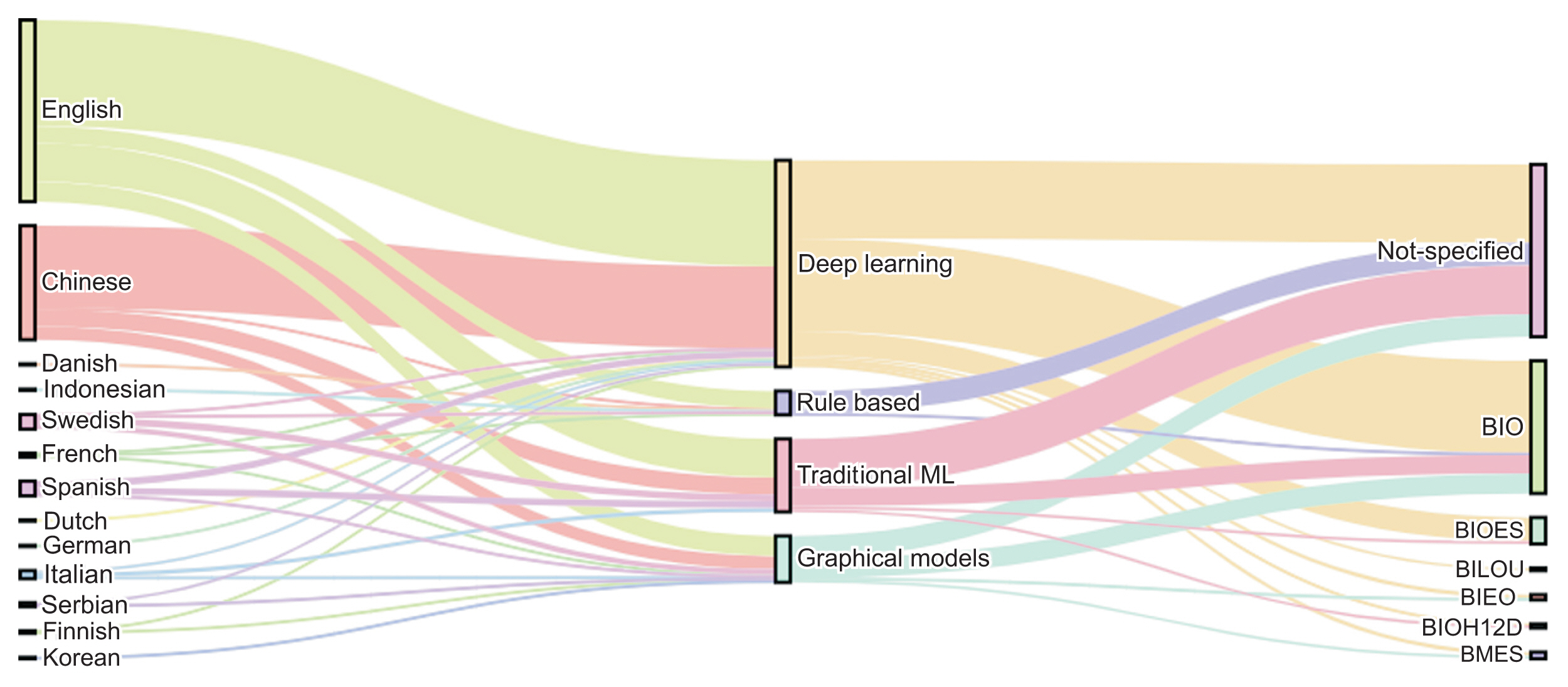
2. Yehia E, Boshnak H, AbdelGaber S, Abdo A, Elzanfaly DS.. Ontology-based clinical information extraction from physician’s free-text notes. J Biomed Inform 2019 98:103276.
https://doi.org/10.1016/j.jbi.2019.103276


3. ElDin HG, AbdulRazek M, Abdelshafi M, Sahlol AT.. Med-Flair: medical named entity recognition for diseases and medications based on Flair embedding. Procedia Comput Sci 2021 189:67-75.
https://doi.org/10.1016/j.procs.2021.05.078

4. Kaplar A, Stosovic M, Kaplar A, Brkovic V, Naumovic R, Kovacevic A.. Evaluation of clinical named entity recognition methods for Serbian electronic health records. Int J Med Inform 2022 164:104805.
https://doi.org/10.1016/j.ijmedinf.2022.104805


5. Neuraz A, Looten V, Rance B, Daniel N, Garcelon N, Llanos LC, et al. Do you need embeddings trained on a massive specialized corpus for your clinical natural language processing task? Stud Health Technol Inform 2019 264:1558-9.
https://doi.org/10.3233/shti190533


6. Kong J, Zhang L, Jiang M, Liu T.. Incorporating multi-level CNN and attention mechanism for Chinese clinical named entity recognition. J Biomed Inform 2021 116:103737.
https://doi.org/10.1016/j.jbi.2021.103737


11. Casillas A, Perez A, Oronoz M, Gojenola K, Santiso S.. Learning to extract adverse drug reaction events from electronic health records in Spanish. Expert Syst Appl 2016 61(1):235-45.
https://doi.org/10.1016/j.eswa.2016.05.034

12. Henriksson A, Kvist M, Dalianis H, Duneld M.. Identifying adverse drug event information in clinical notes with distributional semantic representations of context. J Biomed Inform 2015 57:333-49.
https://doi.org/10.1016/j.jbi.2015.08.013


18. Jung K, LePendu P, Iyer S, Bauer-Mehren A, Percha B, Shah NH.. Functional evaluation of out-of-the-box text-mining tools for data-mining tasks. J Am Med Inform Assoc 2015 22(1):121-31.
https://doi.org/10.1136/amiajnl-2014-002902


21. Wang H, Zhang W, Zeng Q, Li Z, Feng K, Liu L.. Extracting important information from Chinese Operation Notes with natural language processing methods. J Biomed Inform 2014 48:130-6.
https://doi.org/10.1016/j.jbi.2013.12.017


24. Xiong Y, Peng H, Xiang Y, Wong KC, Chen Q, Yan J, et al. Leveraging Multi-source knowledge for Chinese clinical named entity recognition via relational graph convolutional network. J Biomed Inform 2022 128:104035.
https://doi.org/10.1016/j.jbi.2022.104035


27. Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF models for sequence tagging [Internet]. Ithaca (NY): arXiv. org; 2015 [cited at 2023 Sep 30]. Available from:
https://arxiv.org/abs/1508.01991
28. Zhang X, Zhang Y, Zhang Q, Ren Y, Qiu T, Ma J, et al. Extracting comprehensive clinical information for breast cancer using deep learning methods. Int J Med Inform 2019 132:103985.
https://doi.org/10.1016/j.ijmedinf.2019.103985


30. Li PL, Yuan ZM, Tu WN, Yu K, Lu DX.. Medical knowledge extraction and analysis from electronic medical records using deep learning. Chin Med Sci J 2019 34(2):133-9.
https://doi.org/10.24920/003589


31. Suarez-Paniagua V, Rivera Zavala RM, Segura-Bedmar I, Martinez P.. A two-stage deep learning approach for extracting entities and relationships from medical texts. J Biomed Inform 2019 99:103285.
https://doi.org/10.1016/j.jbi.2019.103285


34. Wang Q, Zhou Y, Ruan T, Gao D, Xia Y, He P.. Incorporating dictionaries into deep neural networks for the Chinese clinical named entity recognition. J Biomed Inform 2019 92:103133.
https://doi.org/10.1016/j.jbi.2019.103133


35. Yin M, Mou C, Xiong K, Ren J.. Chinese clinical named entity recognition with radical-level feature and selfattention mechanism. J Biomed Inform 2019 98:103289.
https://doi.org/10.1016/j.jbi.2019.103289


39. Casillas A, Ezeiza N, Goenaga I, Perez A, Soto X.. Measuring the effect of different types of unsupervised word representations on Medical Named Entity Recognition. Int J Med Inform 2019 129:100-6.
https://doi.org/10.1016/j.ijmedinf.2019.05.022


40. Chen Y, Zhou C, Li T, Wu H, Zhao X, Ye K, et al. Named entity recognition from Chinese adverse drug event reports with lexical feature based BiLSTM-CRF and tri-training. J Biomed Inform 2019 96:103252.
https://doi.org/10.1016/j.jbi.2019.103252


41. Liu X, Zhou Y, Wang Z.. Recognition and extraction of named entities in online medical diagnosis data based on a deep neural network. J Vis Commun Image Represent 2019 60:1-15.
https://doi.org/10.1016/j.jvcir.2019.02.001

42. Li Z, Yang J, Gou X, Qi X.. Recurrent neural networks with segment attention and entity description for relation extraction from clinical texts. Artif Intell Med 2019 97:9-18.
https://doi.org/10.1016/j.artmed.2019.04.003


44. Su J, Hu J, Jiang J, Xie J, Yang Y, He B, et al. Extraction of risk factors for cardiovascular diseases from Chinese electronic medical records. Comput Methods Programs Biomed 2019 172:1-10.
https://doi.org/10.1016/j.cmpb.2019.01.007


49. Harnoune A, Rhanoui M, Mikram M, Yousfi S, Elkaimbillah Z, El Asri B.. BERT based clinical knowledge extraction for biomedical knowledge graph construction and analysis. Comput Methods Programs Biomed Update 2021 1:100042.
https://doi.org/10.1016/j.cmpbup.2021.100042

50. Narayanan S, Achan P, Rangan PV, Rajan SP.. Unified concept and assertion detection using contextual multitask learning in a clinical decision support system. J Biomed Inform 2021 122:103898.
https://doi.org/10.1016/j.jbi.2021.103898


51. Thieu T, Maldonado JC, Ho PS, Ding M, Marr A, Brandt D, et al. A comprehensive study of mobility functioning information in clinical notes: entity hierarchy, corpus annotation, and sequence labeling. Int J Med Inform 2021 147:104351.
https://doi.org/10.1016/j.ijmedinf.2020.104351liu


54. El-allaly ED, Sarrouti M, En-Nahnahi N, El Alaoui SO.. MTTLADE: a multi-task transfer learning-based method for adverse drug events extraction. Inf Process Manag 2021 58(3):102473.
https://doi.org/10.1016/j.ipm.2020.102473

55. Uronen L, Salantera S, Hakala K, Hartiala J, Moen H.. Combining supervised and unsupervised named entity recognition to detect psychosocial risk factors in occupational health checks. Int J Med Inform 2022 160:104695.
https://doi.org/10.1016/j.ijmedinf.2022.104695


56. Xiong Y, Peng W, Chen Q, Huang Z, Tang B.. A unified machine reading comprehension framework for cohort selection. IEEE J Biomed Health Inform 2022 26(1):379-87.
https://doi.org/10.1109/jbhi.2021.3095478


57. Zhang B, Liu K, Wang H, Li M, Pan J.. Chinese named-entity recognition via self-attention mechanism and position-aware influence propagation embedding. Data Knowl Eng 2022 139:101983.
https://doi.org/10.1016/j.datak.2022.101983

60. Gerardin C, Wajsburt P, Vaillant P, Bellamine A, Carrat F, Tannier X.. Multilabel classification of medical concepts for patient clinical profile identification. Artif Intell Med 2022 128:102311.
https://doi.org/10.1016/j.artmed.2022.102311


61. Madan S, Julius Zimmer F, Balabin H, Schaaf S, Frohlich H, Fluck J, et al. Deep learning-based detection of psychiatric attributes from German mental health records. Int J Med Inform 2022 161:104724.
https://doi.org/10.1016/j.ijmedinf.2022.104724


62. Narayanan S, Madhuri SS, Ramesh MV, Rangan PV, Rajan SP.. Deep contextual multi-task feature fusion for enhanced concept, negation and speculation detection from clinical notes. Informatics Med Unlocked 2022 34:101109.
https://doi.org/10.1016/j.imu.2022.101109

65. Narayanan S, Mannam K, Achan P, Ramesh MV, Rangan PV, Rajan SP.. A contextual multi-task neural approach to medication and adverse events identification from clinical text. J Biomed Inform 2022 125:103960.
https://doi.org/10.1016/j.jbi.2021.103960


66. El-Allaly ED, Sarrouti M, En-Nahnahi N, Ouatik El Alaoui S.. An attentive joint model with transformer-based weighted graph convolutional network for extracting adverse drug event relation. J Biomed Inform 2022 125:103968.
https://doi.org/10.1016/j.jbi.2021.103968


72. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst 2017;30:5999-6009.
73. Xu Y, Wang Y, Liu T, Liu J, Fan Y, Qian Y, et al. Joint segmentation and named entity recognition using dual decomposition in Chinese discharge summaries. J Am Med Inform Assoc 2014 21(e1):e84-92.
https://doi.org/10.1136/amiajnl-2013-001806


74. Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020 36(4):1234-40.
https://doi.org/10.48550/arXiv.1901.08746


75. Peng Y, Yan S, Lu Z. Transfer learning in biomedical natural language processing: an evaluation of BERT and ELMo on ten benchmarking datasets [Internet]. Ithaca (NY): arXiv.org; 2019 [cited at 2023 Sep 30]. Available from:
https://arxiv.org/abs/1906.05474
76. Ji B, Li S, Yu J, Ma J, Tang J, Wu Q, et al. Research on Chinese medical named entity recognition based on collaborative cooperation of multiple neural network models. J Biomed Inform 2020;104:103395.


77. Sterckx L, Vandewiele G, Dehaene I, Janssens O, Ongenae F, De Backere F, et al. Clinical information extraction for preterm birth risk prediction. J Biomed Inform 2020 110:103544.
https://doi.org/10.1016/j.jbi.2020.103395


78. Kormilitzin A, Vaci N, Liu Q, Nevado-Holgado A.. Med7: a transferable clinical natural language processing model for electronic health records. Artif Intell Med 2021 118:102086.
https://doi.org/10.1016/j.artmed.2021.102086


79. Wei Q, Ji Z, Li Z, Du J, Wang J, Xu J, et al. A study of deep learning approaches for medication and adverse drug event extraction from clinical text. J Am Med Inform Assoc 2020 27(1):13-21.
https://doi.org/10.1093/jamia/ocz063


81. Lin WC, Chen JS, Kaluzny J, Chen A, Chiang MF, Hribar MR.. Extraction of active medications and adherence using natural language processing for glaucoma patients. AMIA Annu Symp Proc 2021;2021:773-82.

83. Chen L, Li Y, Chen W, Liu X, Yu Z, Zhang S.. Utilizing soft constraints to enhance medical relation extraction from the history of present illness in electronic medical records. J Biomed Inform 2018;87:108-17.


84. Dai HJ, Su CH, Wu CS.. Adverse drug event and medication extraction in electronic health records via a cascading architecture with different sequence labeling models and word embeddings. J Am Med Inform Assoc 2020 27(1):47-55.
https://doi.org/10.1093/jamia/ocz120


85. Yang X, Bian J, Fang R, Bjarnadottir RI, Hogan WR, Wu Y.. Identifying relations of medications with adverse drug events using recurrent convolutional neural networks and gradient boosting. J Am Med Inform Assoc 2020 27(1):65-72.
https://doi.org/10.1093/jamia/ocz144


88. Cheng Y, Anick P, Hong P, Xue N.. Temporal relation discovery between events and temporal expressions identified in clinical narrative. J Biomed Inform 2013 46 (Suppl):S48-53.
https://doi.org/10.1016/j.jbi.2013.09.010


90. Kochmar E, Andersen O, Briscoe T. HOO 2012 error recognition and correction shared task: Cambridge University submission report. Proceedings of the 7th Workshop on Innovative Use of NLP for Building Educational Applications; 2012 Jun 7. Montreal, Canada; p. 242-50.
https://doi.org/10.17863/CAM.9671

91. Zhao S, Cai Z, Chen H, Wang Y, Liu F, Liu A.. Adversarial training based lattice LSTM for Chinese clinical named entity recognition. J Biomed Inform 2019 99:103290.
https://doi.org/10.1016/j.jbi.2019.103290


92. Yu G, Yang Y, Wang X, Zhen H, He G, Li Z, et al. Adversarial active learning for the identification of medical concepts and annotation inconsistency. J Biomed Inform 2020 108:103481.
https://doi.org/10.1016/j.jbi.2020.103481


93. Wang C, Wang H, Zhuang H, Li W, Han S, Zhang H, et al. Chinese medical named entity recognition based on multi-granularity semantic dictionary and multimodal tree. J Biomed Inform 2020 111:103583.
https://doi.org/10.1016/j.jbi.2020.103583


97. Hu Y, Ameer I, Zuo W, Peng X, Zhou Y, Li Z, et al. Zeroshot clinical entity recognition using ChatGPT [Internet]. Ithaca (NY): arXiv.org; 2023 [cited at 2023 Sep 30]. Available from:
https://arxiv.org/abs/2303.16416
98. Li X, Zhu X, Ma Z, Liu X, Shas S. Are ChatGPT and GPT-4 general-purpose solvers for financial text analytics? An examination on several typical tasks [Internet]. Ithaca (NY): arXiv.org; 2023 [cited at 2023 Sep 30]. Available from:
https://arxiv.org/abs/2305.05862
99. Lai VD, Ngo NT, Veyseh AP, Man H, Dernoncourt F, Bui T, et al. ChatGPT beyond English: towards a comprehensive evaluation of large language models in multilingual learning [Internet]. Ithaca (NY): arXiv.org; 2023 [cited at 2023 Sep 30]. Available from:
https://arxiv.org/abs/2304.05613